R for text data
Overview
Teaching: FIXME min
Exercises: FIXME minQuestions
FIXME
Objectives
FIXME
Why do this in R?
- Data is rarely clean and tidy
- Misspellings
- White space
- Multiple variables per column
- Inconsistent coding
- Fixing it by hand takes forever
Types of text data
Up until now, we’ve largely treated all text data the same as either all factors or all strings. However, the type of a text column in a tibble determines what you can do with the data. If you want to clean up misspellings or look for patterns in unstructured data, you can do that in a string column. If you want to subset based on a catagory or combine categories, factors are more useful.
This lesson will cover packages that make working with text data easier: stringr and forcats.
These packages are part of the tidyverse, meaning that they work well with dplyr, specifically
the mutate function. We will also cover options in the read_csv function that will allow you to
choose what type the data are when they are imported.
Download Data
Factors
Factors are categorical vectors in R. While some of the operations you can do on them are the same as with character vectors, others differ. They also different in their underlying structure. Character vectors are stored as the characters in each vector. Factors assign a value to each category and then store the values instead of the characters for each item. Given that this reduces the size of your data set, many functions may run faster when categories are set as factors instead of characters.
The data
We will be using a messier version of the surveys data that were used in the dplyr and ggplot2 lessons.
Importing the data
Let’s start by loading the libraries and importing the data with read_csv.
library(tidyverse)
# OR
library(stringr)
library(forcats)
library(ggplot2)
library(dplyr)
library(readr)
library(tidyr)
surveys<-read_csv(file = "data/Portal_rodents_19772002_scinameUUIDs.csv")
Warning: One or more parsing issues, call `problems()` on your data frame for details,
e.g.:
dat <- vroom(...)
problems(dat)
Rows: 35549 Columns: 39
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (22): survey_id, plot_id, species, scientificName, locality, JSON, count...
dbl (16): recordID, mo, dy, yr, period, plot, note1, stake, decimalLatitude,...
lgl (1): note3
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Note that there are a few parsing errors. This error happens becaues read_csv
looks at the first 1000 rows of each column and guess which type that column
should be based on those entries. In our case there are a few entries at the
bottom of the notes columns which don’t fit the type it guessed based on the first
1000 rows. We will add the guess_max argument to have read_csv check the
whole column before it automatically chooses a type for that column.
surveys <- read_csv(file = "data/Portal_rodents_19772002_scinameUUIDs.csv",
guess_max = 40000)
Rows: 35549 Columns: 39
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (25): survey_id, plot_id, species, scientificName, locality, JSON, count...
dbl (14): recordID, mo, dy, yr, period, plot, note1, stake, decimalLatitude,...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Because we imported the data using read_csv, all of the non-numeric columns were converted to the
character class. If we used read.csv, they would all be factors.
Challenge 1
Look at the data columns in the surveys dataset. Which columns should be converted to factors? Which should stay as text? Why?
Hint: should any numeric columns be factors?
Solution to Challenge 1
Changing column classes
#Create a text vector
species<-c("AB", "AS", "AS", "AB")
class(species)
[1] "character"
#convert it to factor
species<-as_factor(species)
species
[1] AB AS AS AB
Levels: AB AS
class(species)
[1] "factor"
#convert back to character
species<-as.character(species)
class(species)
[1] "character"
surveys<- surveys%>%
mutate(species = as_factor(species))
OR
surveys$scientificName <- as_factor(surveys$scientificName)
Or, you could specify the types of all of your columns upon reading.
surveys<-read_csv(file = "data/Portal_rodents_19772002_scinameUUIDs.csv",
col_types = cols(col_character(), #survey_id
col_character(), #recordID
col_integer(), #Month
col_integer(), #day
col_integer(), #year
col_double(), #period
col_factor(), #plot_id
col_factor(), #plot
col_character(), #note1
col_character(), #stake
col_factor(), #species
col_factor(), #scientificName
col_character(), #locality
col_character(), #JSON
col_double(), #decimalLatitude
col_double(), #decimalLongitude
col_factor(), #county
col_factor(), #state
col_factor(), #country
col_factor(), #sex
col_factor(), #age
col_character(), #reprod
col_character(), #testes
col_character(), #vagina
col_character(), #pregnant
col_character(), #nippples
col_character(), #lactation
col_double(), #hfl
col_double(), #wgt
col_character(), #tag
col_character(), #note2
col_character(), #ltag
col_character(), #note3
col_character(), #prevrt
col_integer(), #prevlet
col_character(), #nestdir
col_integer(), #neststk
col_character(), #note4
col_character() #note5
)
)
Challenge 2
Convert the columns you identified in Challenge 1 to factors
Solution to Challenge 2
Fun with Factors
- Recoding factors,
fct_recode() - Reordering factors,
fct_relevel()
Recoding factors
One common function we may need to perform is recoding the factors. In this case we may want to use the month names, instead of their numbers.
surveys$mo_abbv <- surveys$mo %>% as.factor() %>%
fct_recode(Jan='1', Feb='2', Mar='3', Apr='4', May='5',
Jun='6', Jul='7', Aug='8', Sep='9', Oct='10',
Nov='11', Dec='12')
head(surveys)
# A tibble: 6 × 40
survey_id recor…¹ mo dy yr period plot_id plot note1 stake species
<chr> <chr> <int> <int> <int> <dbl> <fct> <fct> <chr> <chr> <fct>
1 491ec41b-0… 6545 9 18 1982 62 4dc160… 13 13 36 AB
2 f280bade-4… 5220 1 24 1982 54 dcbbd3… 20 13 27 AB
3 2b1b4a8a-c… 18932 8 7 1991 162 1e87b1… 19 13 33 AS
4 e98e66c4-5… 20588 1 24 1993 179 91829d… 12 13 41 AS
5 768cdd0d-9… 7020 11 21 1982 63 f24f2d… 24 13 72 AH
6 13851c71-0… 7645 4 16 1983 67 f24f2d… 24 13 21 AH
# … with 29 more variables: scientificName <fct>, locality <chr>, JSON <chr>,
# decimalLatitude <dbl>, decimalLongitude <dbl>, county <fct>, state <fct>,
# country <fct>, sex <fct>, age <fct>, reprod <chr>, testes <chr>,
# vagina <chr>, pregnant <chr>, nipples <chr>, lactation <chr>, hfl <dbl>,
# wgt <dbl>, tag <chr>, note2 <chr>, ltag <chr>, note3 <chr>, prevrt <chr>,
# prevlet <int>, nestdir <chr>, neststk <int>, note4 <chr>, note5 <chr>,
# mo_abbv <fct>, and abbreviated variable name ¹recordID
Easier way to do this.
Getting the month abbreviations recoded more easily. First let’s look at the first 6 months.
surveys$mo %>% head()
[1] 9 1 8 1 11 4
Now we can use the month.abb[] to get back the abbreviated names.
(Still looking at only the first 6)
month.abb[surveys$mo] %>% head()
[1] "Sep" "Jan" "Aug" "Jan" "Nov" "Apr"
Challenge
Add a new column called
mo_fullonto thesurveysdata from that includes the full month name.Shortcut hint: Check out what
month.name[]does.Solution to Challenge
surveys$mo_full <- surveys$mo %>% as.factor() %>% fct_recode(January='1', Febuary='2', March='3', April='4', May='5', June='6', July='7', August='8', September='9', October='10', November='11', December='12')OR
surveys$mo_full <- month.name[surveys$mo]
Reorder factors
If we use the ggplot skills we learned in the last session.
We see that the factors for plot_type display in the order of their
levels, which are in alphabetical order by default.
levels(surveys$plot)
[1] "13" "20" "19" "12" "24" "15" "9" "1" "3" "14" "2" "16" "23" "7" "18"
[16] "22" "8" "10" "17" "21" "6" "4" "11" "5"
surveys %>% filter(!is.na(hfl)) %>%
ggplot(aes(x=plot, y=hfl)) +
geom_boxplot()
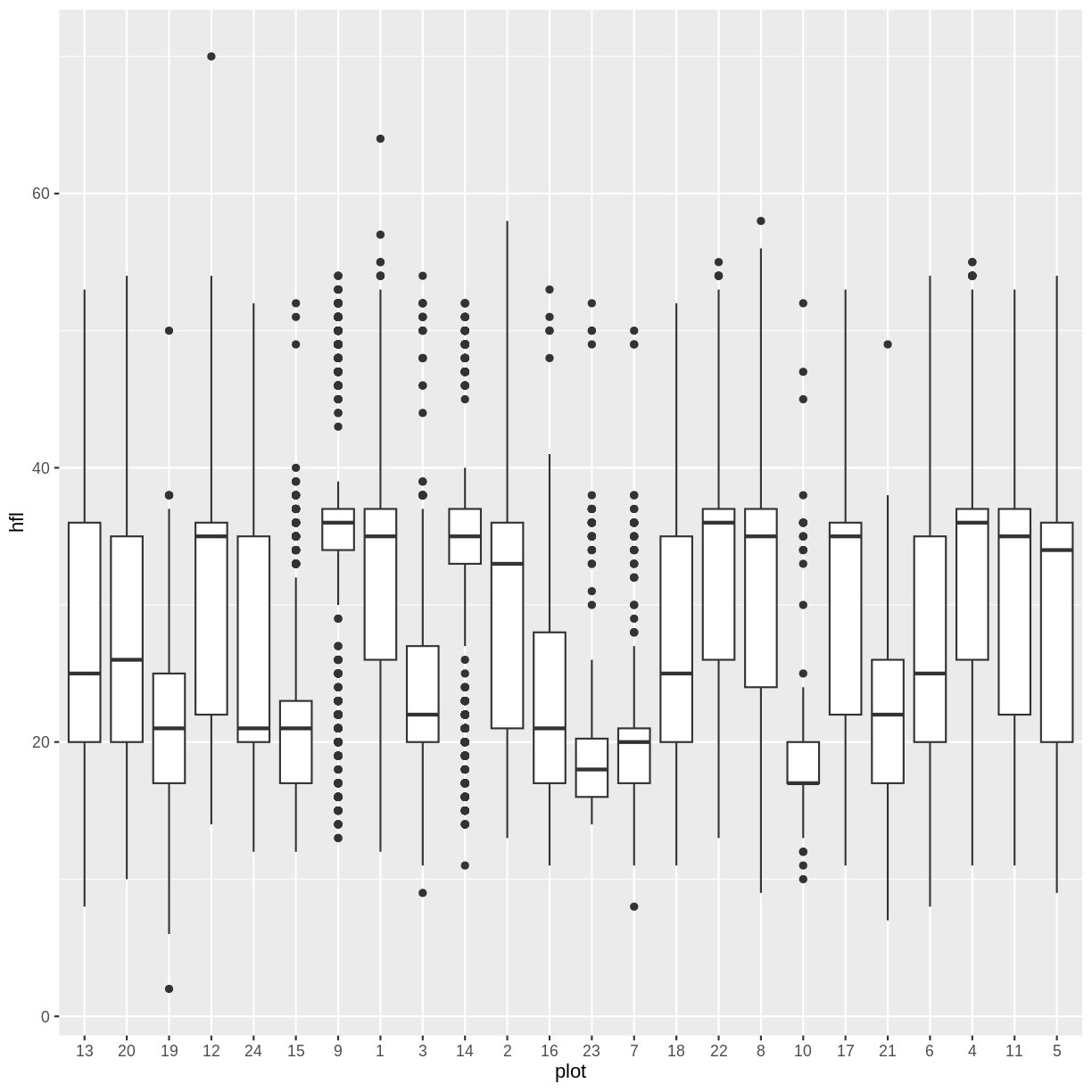
Ordered by number and left pad
Let’s put the plots in order by their number using the fct_relevel function.
surveys$plot %>%
levels() %>%
sort()
[1] "1" "10" "11" "12" "13" "14" "15" "16" "17" "18" "19" "2" "20" "21" "22"
[16] "23" "24" "3" "4" "5" "6" "7" "8" "9"
This sort is sorting alphabetically and by place. To fix the sorting, we can add a leading zero and ‘left pad’ the names using a string method.
str_pad(surveys$plot, width = 2, side = "left", pad="0") %>% head(10)
[1] "13" "20" "19" "12" "24" "24" "15" "09" "15" "13"
surveys$plot <- str_pad(surveys$plot, width = 2, side = "left", pad="0") %>% as_factor()
order <- surveys$plot %>%
levels() %>%
sort()
surveys$plot <- fct_relevel(surveys$plot, order)
levels(surveys$plot)
[1] "01" "02" "03" "04" "05" "06" "07" "08" "09" "10" "11" "12" "13" "14" "15"
[16] "16" "17" "18" "19" "20" "21" "22" "23" "24"
surveys %>% filter(!is.na(hfl)) %>%
ggplot(aes(x=plot, y=hfl)) +
geom_boxplot()
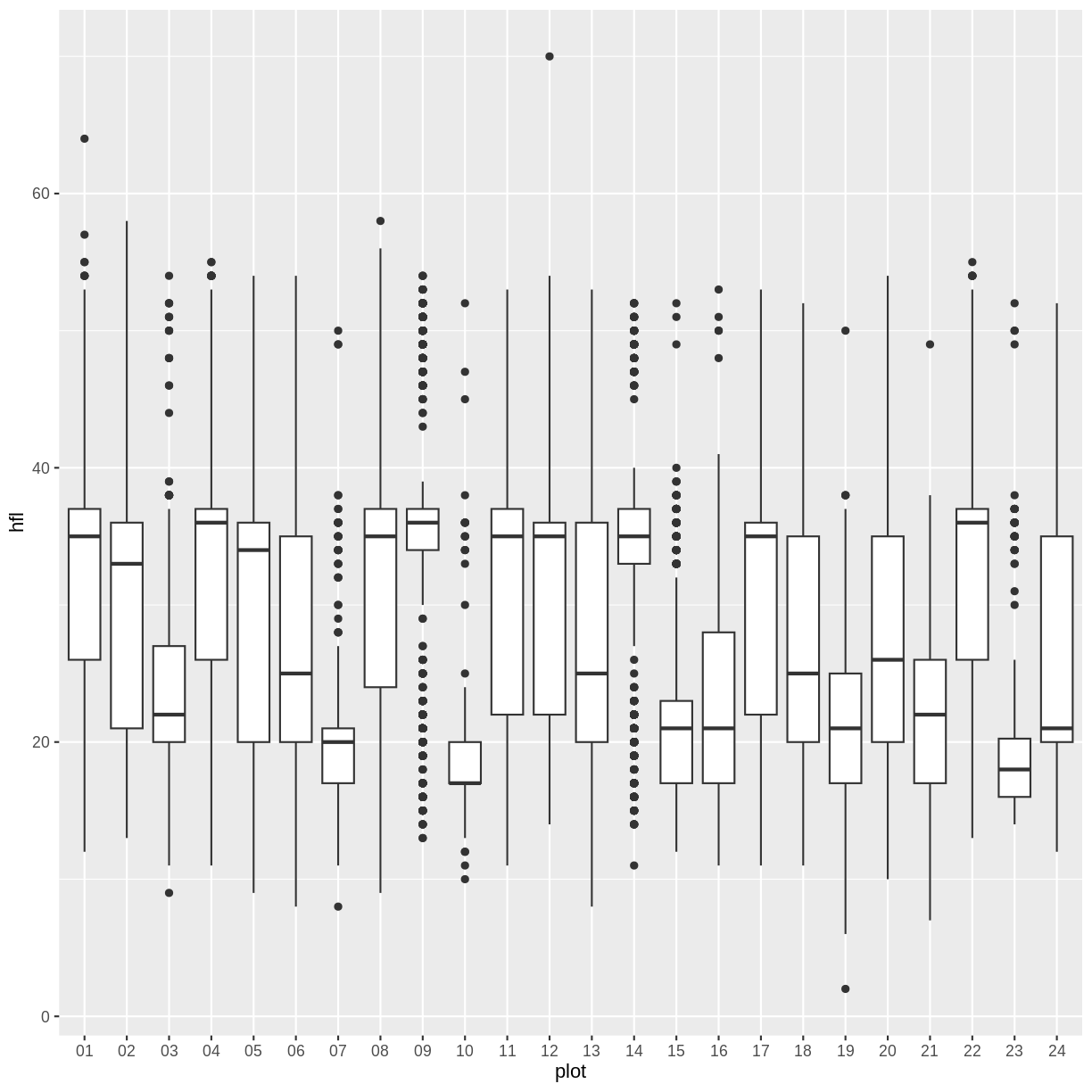
We can also reorder only a subset of the levels without having to specify
all of the levels by using the after= argument
We can say 1 (after the first level) to Inf (after everything) instead of
typing out each of the levels in order.
We know from other information that the levels ‘2’, ‘4’, ‘8’, ‘11’, ‘12’, ‘17’, ‘22’ are the control plots. Let’s try putting the level ‘2’ at the end so we can see all the controls to the right.
surveys$plot <- surveys$plot %>% fct_relevel('2', after= Inf)
Warning: 1 unknown level in `f`: 2
Now if we plot the same box plot above, plot 2 is now on the far right. You can this to reorder the categories in other plots as well.
surveys %>%
filter(!is.na(hfl)) %>%
ggplot(aes(x=plot, y=hfl)) +
geom_boxplot()
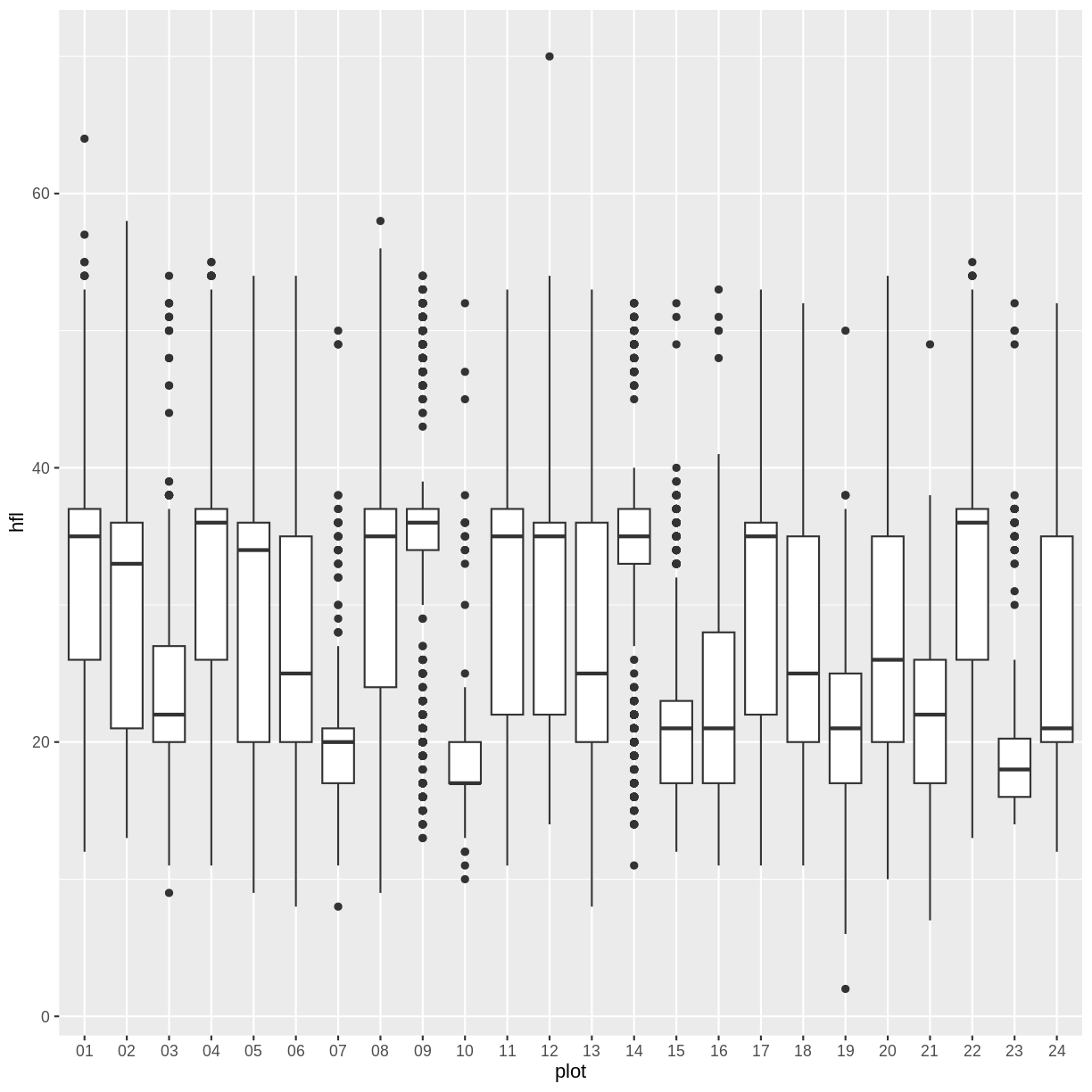
Challenge
Reorder the
plot’s in the boxplot above so all the control plots are on the right.Solution to Challenge
surveys$plot<- surveys$plot %>% fct_relevel('2', '4', '8', '11', '12', '17', '22', after= Inf) surveys %>% filter(!is.na(hfl)) %>% ggplot(aes(x=plot, y=hfl)) + geom_boxplot()
Cleaning up text data
When text data is entered by hand, small differences can be introduced that
aren’t easy to see with the human eye, but are important to the computer.
One easy way to identify these small differences is the count function.
surveys%>%
count(scientificName)
# A tibble: 27 × 2
scientificName n
<fct> <int>
1 Amphispiza bilineata 291
2 Ammodramus savannarum 2
3 Ammospermophilis harrisi 1
4 Ammospermophilus harrisi 435
5 Ammospermophilus harrisii 1
6 Amphespiza bilineata 7
7 Amphispiza bilineatus 1
8 Amphispiza cilineata 1
9 Amphispizo bilineata 1
10 Baiomys taylori 46
# … with 17 more rows
You can see some very similar species names, for example: “Ammospermophilis harrisi”, “Ammospermophilus harrisi”, “Ammospermophilus harrisii”. However one spelling has many more records than the others. How can we fix the spellings?
surveys$scientificName <- fct_explicit_na(surveys$scientificName)
surveys$scientificName <- fct_collapse(surveys$scientificName,
"Ammospermophilus harrisi"=c("Ammospermophilus harrisi",
"Ammospermophilis harrisi",
"Ammospermophilus harrisii"),
"Amphespiza bilineata" = c("Amphispiza bilineatus",
"Amphispiza cilineata",
"Amphispizo bilineata"))
We can see the change by looking at the count again.
surveys%>%
count(scientificName)
# A tibble: 22 × 2
scientificName n
<fct> <int>
1 Amphispiza bilineata 291
2 Ammodramus savannarum 2
3 Ammospermophilus harrisi 437
4 Amphespiza bilineata 10
5 Baiomys taylori 46
6 Calamospiza melanocorys 1
7 Callipepla squamata 1
8 Campylorhynchus brunneicapillus 1
9 Chaetodipus baileyi 2
10 Cnemidophorus tigris 1
# … with 12 more rows
Challenge
- Find all the possible variants on the country name “United States””
- Change them all to the most common variant.
Solution to Challenge
surveys %>% count(country) # We can see that "United States of America", "UNITED STATES", and "US" # Are possible options with "UNITED STATES" being the most common. surveys$country <- fct_collapse(surveys$country, "UNITED STATES" = c("United States of America", "US"))
Splitting Variables
Next we may want to split the scientific names into genus and species columns as we have seen in the cleaned version of the data.
surveys <- separate(surveys, scientificName, c("genusName", "speciesName"), sep="\\s", remove = FALSE)
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16923
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16924
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16925
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16926
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16927
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16928
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16929
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16930
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16931
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16932
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16933
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16934
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16935
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16936
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16937
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16938
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16939
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16940
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16941
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16942
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16943
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16944
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16945
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16946
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16947
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16948
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16949
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16950
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16951
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16952
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16953
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16954
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16955
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16956
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16957
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16958
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16959
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16960
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16961
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 16962
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20220
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20221
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20222
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20223
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20224
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20225
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20226
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20227
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20228
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20229
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20230
Warning in gregexpr(pattern, x, perl = TRUE): PCRE error
'UTF-8 error: isolated byte with 0x80 bit set'
for element 20231
Warning: Expected 2 pieces. Missing pieces filled with `NA` in 15370 rows [16923, 16924,
16925, 16926, 16927, 16928, 16929, 16930, 16931, 16932, 16933, 16934, 16935,
16936, 16937, 16938, 16939, 16940, 16941, 16942, ...].
Joining Variables
In some of our plots we may want to label with the full scientific name. To do so we can add a new column which joins two strings together. Before we get into vectors lets try an example with two strings
name = "Sarah"
str_c("Hi my name is ", name)
[1] "Hi my name is Sarah"
We can similarly use this on vectors. We can make one column that has the latitude and longitude.
surveys$latnlong <- str_c(surveys$decimalLatitude, " ", surveys$decimalLongitude)
Another function that you could have used here is paste()
Other stringr functions
Next, let’s see if all our recordIDs are the same length.
str_length(surveys$recordID) %>% head()
[1] 4 4 5 5 4 4
We can see that they are not all the same length but it is hard
to see what the different lengths are lets see the different
lengths using the unique() function.
str_length(surveys$recordID) %>% unique()
[1] 4 5 1 2 3
Challenge
Use the use stringr function we learned earlier to make all the recordIDs the same length.
Solution to Challenge
surveys$recordID<- surveys$recordID %>% str_pad(width = 5, side = "left", pad = "0")
Another string function we can use is to get a subset of a string.
We can use that function, str_sub() to create abbvs for the genera.
We can then add those abbrvs as their own column
str_sub(surveys$genusName, 1, 5) %>% head()
[1] "Amphi" "Amphi" "Ammod" "Ammod" "Ammos" "Ammos"
surveys <- surveys %>% mutate(genusAbbv = str_sub(surveys$genusName, 1, 5))
Finding patterns
Rstudio Regular expression Cheatsheet Rstudio stingr Cheatsheet
Find the scientific names with punctuation in them.
str_detect(surveys$scientificName, "Dip") %>% head()
[1] FALSE FALSE FALSE FALSE FALSE FALSE
str_detect(surveys$scientificName, "Dip") %>% unique()
[1] FALSE TRUE
str_subset(surveys$scientificName, "Dip") %>% head()
[1] "Dipodomys merriami" "Dipodomys merriami" "Dipodomys merriami"
[4] "Dipodomys merriami" "Dipodomys merriami" "Dipodomys merriami"
str_subset(surveys$scientificName, "Dip") %>% unique()
[1] "Dipodomys merriami" "Dipodomys ordii" "Dipodomys spectabilis"
[4] "Dipodomys�sp."
str_subset(surveys$scientificName, "[[:punct:]]") %>% head()
[1] "Dipodomys�sp." "Dipodomys�sp." "Dipodomys�sp." "Dipodomys�sp."
[5] "Dipodomys�sp." "Dipodomys�sp."
str_subset(surveys$scientificName, "[[:punct:]]") %>% unique()
[1] "Dipodomys�sp." "Onychomys�sp." "(Missing)"
Let’s replace all the puntuation characters with a space for the moment.
statement = "Sarah is the instructor"
str_replace(statement, "a", "e")
[1] "Serah is the instructor"
str_replace_all(statement, "a", "e")
[1] "Sereh is the instructor"
surveys$scientificName <- str_replace_all(surveys$scientificName, "[[:punct:]]", " ")
surveys %>% count(scientificName)
# A tibble: 22 × 2
scientificName n
<chr> <int>
1 " Missing " 15318
2 "Ammodramus savannarum" 2
3 "Ammospermophilus harrisi" 437
4 "Amphespiza bilineata" 10
5 "Amphispiza bilineata" 291
6 "Baiomys taylori" 46
7 "Calamospiza melanocorys" 1
8 "Callipepla squamata" 1
9 "Campylorhynchus brunneicapillus" 1
10 "Chaetodipus baileyi" 2
# … with 12 more rows
Other pattern matching commands that can be useful:
str_match()
str_count()
str_locate()
str_extract()
Remove leading/trailing whitespace
Now we have some extra whitespace to remove from the scientificName column.
We can use the str_trim function
str_subset(surveys$scientificName, "Miss") %>% head()
[1] " Missing " " Missing " " Missing " " Missing " " Missing " " Missing "
str_subset(surveys$scientificName, "Miss")[1] %>% str_trim()
[1] "Missing"
str_trim(surveys$scientificName) %>% str_subset("Miss") %>% head()
[1] "Missing" "Missing" "Missing" "Missing" "Missing" "Missing"
surveys$scientificName <- str_trim(surveys$scientificName)
Write back to a csv file
write_csv(surveys, "cleaned_surveys_20191005_slr.csv")
Key Points
FIXME