Content from Overview of Amazon SageMaker
Last updated on 2024-11-05 | Edit this page
Amazon SageMaker is a comprehensive machine learning platform that empowers users to build, train, tune, and deploy models at scale. Designed to streamline the ML workflow, SageMaker supports data scientists and researchers in tackling complex machine learning problems without needing to manage underlying infrastructure. This allows you to focus on developing and refining your models while leveraging AWS’s robust computing resources for efficient training and deployment.
Why use SageMaker for machine learning?
SageMaker provides several features that make it an ideal choice for researchers and ML practitioners:
End-to-end workflow: SageMaker covers the entire ML pipeline, from data preprocessing to model deployment. This unified environment reduces the need to switch between platforms or tools, enabling users to set up, train, tune, and deploy models seamlessly.
Flexible compute options: SageMaker lets you easily select instance types tailored to your project needs. For compute-intensive tasks, such as training deep learning models, you can switch to GPU instances for faster processing. SageMaker’s scalability also supports parallelized training, enabling you to distribute large training jobs across multiple instances, which can significantly speed up training time for large datasets and complex models.
Efficient hyperparameter tuning: SageMaker provides powerful tools for automated hyperparameter tuning, allowing users to perform complex cross-validation (CV) searches with a single chunk of code. This feature enables you to explore a wide range of parameters and configurations efficiently, helping you find optimal models without manually managing multiple training runs.
Support for Custom Scripts: While SageMaker offers built-in algorithms, it also allows users to bring their own customized scripts. This flexibility is crucial for researchers developing unique models or custom algorithms. SageMaker’s support for Docker containers allows you to deploy fully customized code for training, tuning, and inference on scalable AWS infrastructure.
Cost management and monitoring: SageMaker includes built-in monitoring tools to help you track and manage costs, ensuring you can scale up efficiently without unnecessary expenses. With thoughtful usage, SageMaker can be very affordable—for example, training roughly 100 models on a small dataset (under 1GB) can cost less than $20, making it accessible for many research projects.
SageMaker is designed to support machine learning at any scale, making it a strong choice for projects ranging from small experiments to large research deployments. With robust tools for every step of the ML process, it empowers researchers and practitioners to bring their models from development to production efficiently and effectively.
Content from Data Storage: Setting up S3
Last updated on 2024-11-05 | Edit this page
Overview
Questions
- How can I store and manage data effectively in AWS for SageMaker workflows?
- What are the best practices for using S3 versus EC2 storage for machine learning projects?
Objectives
- Explain data storage options in AWS for machine learning projects.
- Describe the advantages of S3 for large datasets and multi-user workflows.
- Outline steps to set up an S3 bucket and manage data within SageMaker.
Step 1: Data storage
Hackathon Attendees: All data uploaded to AWS must relate to your specific Kaggle challenge, except for auxiliary datasets for transfer learning or pretraining. DO NOT upload any restricted or sensitive data to AWS.
Options for storage: EC2 Instance or S3
When working with SageMaker and other AWS services, you have options for data storage, primarily EC2 instances or S3.
What is an EC2 instance?
An Amazon EC2 (Elastic Compute Cloud) instance is a virtual server environment where you can run applications, process data, and store data temporarily. EC2 instances come in various types and sizes to meet different computing and memory needs, making them versatile for tasks ranging from light web servers to intensive machine learning workloads. In SageMaker, the notebook instance itself is an EC2 instance configured to run Jupyter notebooks, enabling direct data processing.
When to store data directly on EC2
Using an EC2 instance for data storage can be useful for temporary or small datasets, especially during processing within a Jupyter notebook. However, this storage is not persistent; if the instance is stopped or terminated, the data is erased. Therefore, EC2 is ideal for one-off experiments or intermediate steps in data processing.
Limitations of EC2 storage
- Scalability: EC2 storage is limited to the instance’s disk capacity, so it may not be ideal for very large datasets.
- Cost: EC2 storage can be more costly for long-term use compared to S3.
- Data Persistence: EC2 data may be lost if the instance is stopped or terminated, unless using Elastic Block Store (EBS) for persistent storage.
What is an S3 bucket?
Storing data in an S3 bucket is generally preferred
for machine learning workflows on AWS, especially when using SageMaker.
An S3 bucket is a container in Amazon S3 (Simple Storage Service) where
you can store, organize, and manage data files. Buckets act as the
top-level directory within S3 and can hold a virtually unlimited number
of files and folders, making them ideal for storing large datasets,
backups, logs, or any files needed for your project. You access objects
in a bucket via a unique S3 URI (e.g.,
s3://your-bucket-name/your-file.csv), which you can use to
reference data across various AWS services like EC2 and SageMaker.
Benefits of using S3 (recommended for SageMaker and ML workflows)
For flexibility, scalability, and cost efficiency, store data in S3 and load it into EC2 as needed. This setup allows:
- Scalability: S3 handles large datasets efficiently, enabling storage beyond the limits of an EC2 instance’s disk space.
- Cost efficiency: S3 storage costs are generally lower than expanding EC2 disk volumes. You only pay for the storage you use.
- Separation of storage and compute: You can start and stop EC2 instances without losing access to data stored in S3.
- Integration with AWS services: SageMaker can read directly from and write back to S3, making it ideal for AWS-based workflows.
- Easy data sharing: Datasets in S3 are easier to share with team members or across projects compared to EC2 storage.
- Cost-effective data transfer: When S3 and EC2 are in the same region, data transfer between them is free.
Recommended approach: S3 buckets
Hackathon attendees: When you setup your bucket for your actual project, note that you will only need one bucket for your whole team. Team members will have the proper permissions to access buckets on our shared account.
Summary steps to access S3 and upload your dataset
- Log in to AWS Console and navigate to S3.
- Create a new bucket or use an existing one.
- Upload your dataset files.
- Use the object URL to reference your data in future experiments.
Detailed procedure
-
Sign in to the AWS Management Console
- Log in to AWS Console using your credentials.
-
Navigate to S3
- Type “S3” in the search bar
- Protip: select the star icon to save S3 as a bookmark in your AWS toolbar
- Select S3 - Scalable Storage in the Cloud
-
Create a new bucket
- Click Create Bucket and enter a unique name, and
note that bucket name must not contain uppercase characters.
Hackathon participants: Use the following convention
for your bucket name:
teamname_datasetname(e.g.,myawesometeam-titanic). -
Region: Leave as is (likely
us-east-1(US East N. Virginia)) - Access Control: Disable ACLs (recommended).
- Public Access: Turn on “Block all public access”.
- Versioning: Disable unless you need multiple versions of objects.
-
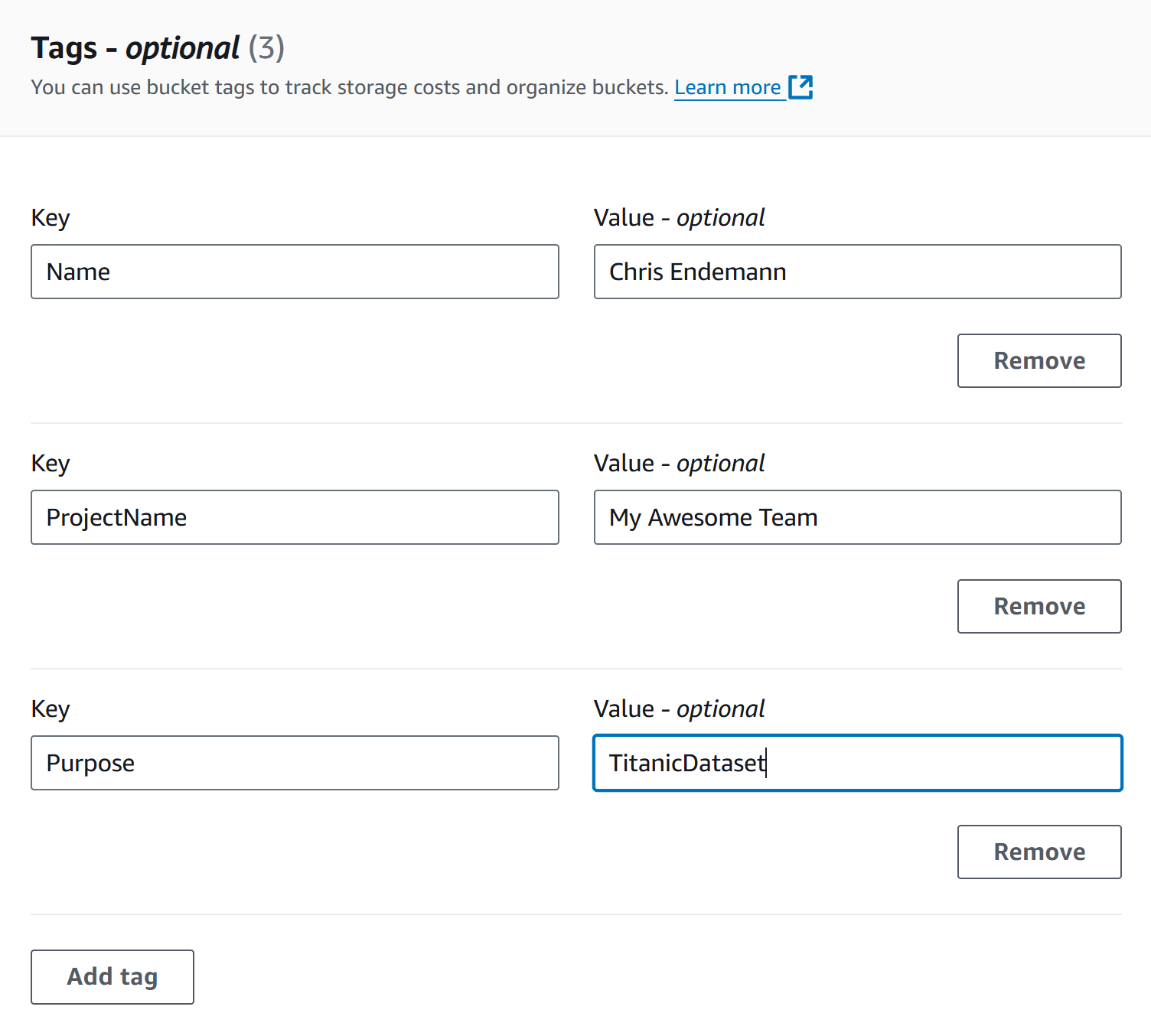
Tags: Adding tags to your S3 buckets is a great way
to track project-specific costs and usage over time, especially as data
and resources scale up. While tags are required for hackathon
participants, we suggest that all users apply tags to easily identify
and analyze costs later. Hackathon participants: Use
the following convention for your bucket name
- Name: Your Name
- ProjectName: Your team’s name
-
Purpose: Dataset name (e.g., titanic if you’re
following along with this workshop)
- Click Create Bucket at the bottom once everything above has been configured
- Click Create Bucket and enter a unique name, and
note that bucket name must not contain uppercase characters.
Hackathon participants: Use the following convention
for your bucket name:
-
Edit bucket policy Once the bucket is created, you’ll be brought to a page that shows all of your current buckets (and those on our shared account). We’ll have to edit our bucket’s policy to allow ourselves proper access to any files stored there (e.g., read from bucket, write to bucket). To set these permissions…
- Click on the name of your bucket to bring up additional options and settings.
- Click the Permissions tab
- Scroll down to Bucket policy and click Edit. Paste the following policy, editing the bucket name “myawesometeam-titanic” to reflect your bucket’s name
JSON
{
"Version": "2012-10-17",
"Statement": [
{
"Effect": "Allow",
"Principal": {
"AWS": "arn:aws:iam::183295408236:role/ml-sagemaker-use"
},
"Action": [
"s3:GetObject",
"s3:PutObject",
"s3:DeleteObject",
"s3:ListMultipartUploadParts"
],
"Resource": [
"arn:aws:s3:::myawesometeam-titanic",
"arn:aws:s3:::myawesometeam-titanic/*"
]
}
]
}For hackathon attendees, this policy grants the
ml-sagemaker-use IAM role access to specific S3 bucket
actions, ensuring they can use the bucket for reading, writing,
deleting, and listing parts during multipart uploads. Attendees should
apply this policy to their buckets to enable SageMaker to operate on
stored data.
General guidance for setting up permissions outside the hackathon
For those not participating in the hackathon, it’s essential to
create a similar IAM role (such as ml-sagemaker-use) with
policies that provide controlled access to S3 resources, ensuring only
the necessary actions are permitted for security and
cost-efficiency.
Create an IAM role: Set up an IAM role for SageMaker to assume, with necessary S3 access permissions, such as
s3:GetObject,s3:PutObject,s3:DeleteObject, ands3:ListMultipartUploadParts, as shown in the policy above.Attach permissions to S3 buckets: Attach bucket policies that specify this role as the principal, as in the hackathon example.
More information: For a detailed guide on setting up roles and policies for SageMaker, refer to the AWS SageMaker documentation on IAM roles and policies. This resource explains role creation, permission setups, and policy best practices tailored for SageMaker’s operations with S3 and other AWS services.
This setup ensures that your SageMaker operations will have the access needed without exposing the bucket to unnecessary permissions or external accounts.
-
Upload files to the bucket
- Navigate to the Objects tab of your bucket, then Upload.
-
Add Files (e.g.,
titanic_train.csv,titanic_test.csv) and click Upload to complete.
-
Take note of S3 URI for your data
- After uploading, click on a file to find its Object
URI (e.g.,
s3://titanic-dataset-test/test.csv). We’ll use this URI to load data into SageMaker later.
- After uploading, click on a file to find its Object
URI (e.g.,
S3 bucket costs
S3 bucket storage incurs costs based on data storage, data transfer, and request counts.
Storage costs
- Storage is charged per GB per month. Typical: Storing 10 GB costs approximately $0.23/month in S3 Standard (us-east-1).
- Pricing Tiers: S3 offers multiple storage classes (Standard, Intelligent-Tiering, Glacier, etc.), with different costs based on access frequency and retrieval times. Standard S3 fits most purposes. If you’re curious about other tiers, refer to AWS’s S3 Pricing Information.
- To calculate specific costs based on your needs, storage class, and region, refer to AWS’s S3 Pricing Information.
Data transfer costs
- Uploading data to S3 is free.
- Downloading data (out of S3) incurs charges (~$0.09/GB). Be sure to take note of this fee, as it can add up fast for large datasets.
- In-region transfer (e.g., S3 to EC2) is free, while cross-region data transfer is charged (~$0.02/GB).
Request costs
- GET requests are $0.0004 per 1,000 requests. In the context of Amazon S3, “GET” requests refer to the action of retrieving or downloading data from an S3 bucket. Each time a file or object is accessed in S3, it incurs a small cost per request. This means that if you have code that reads data from S3 frequently, such as loading datasets repeatedly, each read operation counts as a GET request.
Challenge Exercise: Calculate Your Project’s Data Costs
Estimate the total cost of storing your project data in S3 for one month, using the following dataset sizes and assuming:
- Storage duration: 1 month
- Storage region: us-east-1
- Storage class: S3 Standard
- Data will be retrieved 100 times for model training
(
GETrequests) - Data will be deleted after the project concludes, incurring data retrieval and deletion costs
Dataset sizes to consider:
- 1 GB
- 10 GB
- 100 GB
- 1 TB
Hints
- S3 storage cost: $0.023 per GB per month (us-east-1)
- Data transfer cost (retrieval/deletion): $0.09 per GB (us-east-1 out to internet)
-
GETrequests cost: $0.0004 per 1,000 requests (each model training will incur oneGETrequest)
Check the AWS S3 Pricing page for more details.
Using the S3 Standard rate in us-east-1:
-
1 GB:
- Storage: 1 GB * $0.023 = $0.023
-
Retrieval/Deletion: 1 GB * $0.09 = $0.09
-
GET Requests: 100 requests * $0.0004 per 1,000 =
$0.00004
- Total Cost: $0.11304
-
10 GB:
- Storage: 10 GB * $0.023 = $0.23
-
Retrieval/Deletion: 10 GB * $0.09 = $0.90
-
GET Requests: 100 requests * $0.0004 per 1,000 =
$0.00004
- Total Cost: $1.13004
-
100 GB:
- Storage: 100 GB * $0.023 = $2.30
-
Retrieval/Deletion: 100 GB * $0.09 = $9.00
-
GET Requests: 100 requests * $0.0004 per 1,000 =
$0.00004
- Total Cost: $11.30004
-
1 TB (1024 GB):
- Storage: 1024 GB * $0.023 = $23.55
-
Retrieval/Deletion: 1024 GB * $0.09 = $92.16
-
GET Requests: 100 requests * $0.0004 per 1,000 =
$0.00004
- Total Cost: $115.71004
These costs assume no additional request charges beyond those for
retrieval, storage, and GET requests for training.
Removing unused data
Choose one of these options:
Option 1: Delete data only
- When to Use: You plan to reuse the bucket.
-
Steps:
- Go to S3, navigate to the bucket.
- Select files to delete, then Actions > Delete.
-
CLI (optional):
!aws s3 rm s3://your-bucket-name --recursive
Option 2: Delete the S3 bucket entirely
- When to Use: You no longer need the bucket or data.
-
Steps:
- Select the bucket, click Actions > Delete.
- Type the bucket name to confirm deletion.
Deleting the bucket stops all costs associated with storage, requests, and data transfer.
Key Points
- Use S3 for scalable, cost-effective, and flexible storage.
- EC2 storage is fairly uncommon, but may be suitable for small, temporary datasets.
- Track your S3 storage costs, data transfer, and requests to manage expenses.
- Regularly delete unused data or buckets to avoid ongoing costs.
Content from Notebooks as Controllers
Last updated on 2024-11-06 | Edit this page
Overview
Questions
- How do you set up and use SageMaker notebooks for machine learning tasks?
- How can you manage compute resources efficiently using SageMaker’s controller notebook approach?
Objectives
- Describe how to use SageMaker notebooks for ML workflows.
- Set up a Jupyter notebook instance as a controller to manage compute tasks.
- Use SageMaker SDK to launch training and tuning jobs on scalable instances.
Step 2: Running Python code with SageMaker notebooks
Amazon SageMaker provides a managed environment to simplify the process of building, training, and deploying machine learning models. By using SageMaker, you can focus on model development without needing to manually provision resources or set up environments. In this episode, we’ll guide you through setting up a SageMaker notebook instance—a Jupyter notebook hosted on AWS specifically for running SageMaker jobs. This setup allows you to efficiently manage and monitor machine learning workflows directly from a lightweight notebook controller. We’ll also cover loading data in preparation for model training and tuning in future episodes, using the Titanic dataset stored in S3.
Note for hackathon attendees: We’ll use SageMaker notebook instances (not the full SageMaker Studio environment) for simpler instance management and streamlined resource usage, ideal for collaborative projects or straightforward ML tasks.
Using the notebook as a controller
In this setup, the notebook instance functions as a
controller to manage more resource-intensive compute
tasks. By selecting a minimal instance (e.g., ml.t3.medium)
for the notebook, you can perform lightweight operations and leverage
the SageMaker Python SDK to launch more powerful,
scalable compute instances when needed for model training, batch
processing, or hyperparameter tuning. This approach minimizes costs by
keeping your controller instance lightweight while accessing the full
power of SageMaker for demanding tasks.
Summary of key steps
- Navigate to SageMaker in AWS.
- Create a Jupyter notebook instance as a controller.
- Set up the Python environment within the notebook.
- Load the Titanic dataset from S3.
- Use SageMaker SDK to launch training and tuning jobs on powerful instances (covered in next episodes).
- View and monitor training/tuning progress (covered in next episodes).
Detailed procedure
1. Navigate to SageMaker
- In the AWS Console, search for SageMaker.
- Protip: select the star icon to save SageMaker as a bookmark in your AWS toolbar
- Select SageMaker - Build, Train, and Deploy Models.
2. Create a new notebook instance
- In the SageMaker left-side menu, click on Notebooks, then click Create notebook instance.
-
Notebook name: Enter a name that reflects your
notebook’s primary user (your name), dataset (titanic), purpose
(train-tune), and models utilized (XGBoost-NN). Hackathon
attendees must use the following convention:
TeamName-YourName-Dataset-NotebookPurpose(s)-Model(s) (e.g.,
MyAwesomeTeam-ChrisEndemann-Titanic-Train-Tune-XGBoost-NN). -
Instance type: Start with a small instance type,
such as
ml.t3.medium. You can scale up later as needed for intensive tasks, which will be managed by launching separate training jobs from this notebook. For guidance on common instances for ML procedures, refer to this spreadsheet. - Platform identifier: You can leave this as the default.
-
Permissions and encryption:
-
IAM role: Choose an existing role or create a new
one. Hackathon attendees should select
‘ml-sagemaker-use’. The role should include the
AmazonSageMakerFullAccesspolicy to enable access to AWS services like S3. - Root access: Enable root access to notebook.
- Encryption key (optional): Specify a KMS key for encrypting data at rest if needed. Otherwise, leave it blank.
-
IAM role: Choose an existing role or create a new
one. Hackathon attendees should select
‘ml-sagemaker-use’. The role should include the
- Network (optional): Networking settings are optional. Configure them if you’re working within a specific VPC or need network customization.
- Git repositories configuration (optional): You don’t need to complete this configuration. Instead, we’ll run a clone command from our notebook later to get our repo setup. This approach is a common strategy (allowing some flexiblity in which repo you use for the notebook.
- Tags (required for hackathon attendees): Adding tags helps track and organize resources for billing and management. This is particularly useful when you need to break down expenses by project, task, or team. Please use the tags found in the below image to track your notebook’s resource usage.
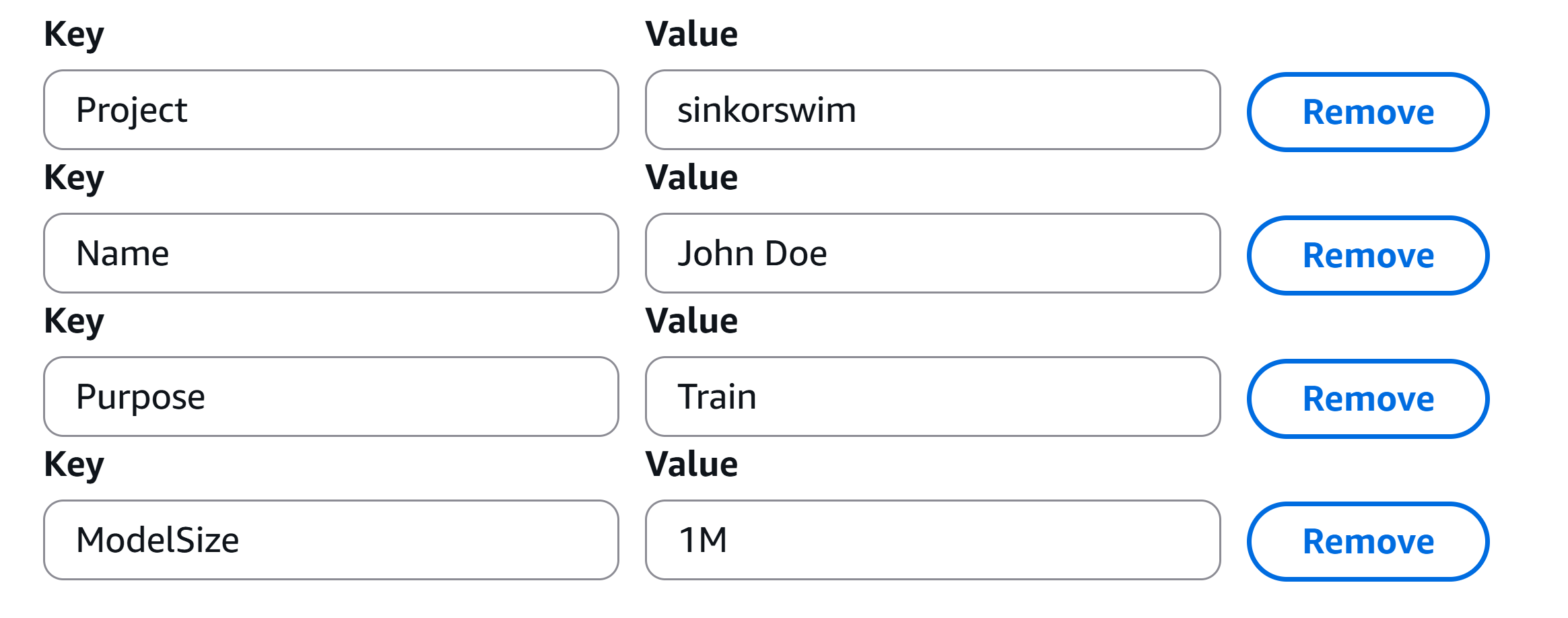
- Click Create notebook instance. It may take a few minutes for the instance to start. Once its status is InService, you can open the notebook instance and start coding.
Managing training and tuning with the controller notebook
In the next couple expisodes, we’ll use the SageMaker Python SDK within the notebook to launch compute-heavy tasks on more powerful instances as needed. Examples of tasks to launch include:
-
Training a model: Use the SDK to submit a training
job, specifying a higher-powered instance (e.g.,
ml.p2.xlargeorml.m5.4xlarge) based on your model’s resource requirements. - Hyperparameter tuning: Configure and launch tuning jobs, allowing SageMaker to automatically manage multiple powerful instances for optimal tuning.
This setup allows you to control costs by keeping the notebook instance minimal and only incurring costs for larger instances when they are actively training or tuning models. Detailed guidance on training, tuning, and batch processing will follow in later episodes.
For more details, refer to the SageMaker Python SDK documentation for example code on launching and managing remote training jobs.
Key Points
- Use a minimal SageMaker notebook instance as a controller to manage larger, resource-intensive tasks.
- Launch training and tuning jobs on scalable instances using the SageMaker SDK.
- Tags can help track costs effectively, especially in multi-project or team settings.
- Use the SageMaker SDK documentation to explore additional options for managing compute resources in AWS.
Content from Accessing and Managing Data in S3 with SageMaker Notebooks
Last updated on 2024-11-14 | Edit this page
Overview
Questions
- How can I load data from S3 into a SageMaker notebook?
- How do I monitor storage usage and costs for my S3 bucket?
- What steps are involved in pushing new data back to S3 from a notebook?
Objectives
- Read data directly from an S3 bucket into memory in a SageMaker notebook.
- Check storage usage and estimate costs for data in an S3 bucket.
- Upload new files from the SageMaker environment back to the S3 bucket.
Initial setup
Open .ipynb notebook
Once your newly created notebook instance (“SageMaker
notebook”) shows as InService, open the instance in Jupyter
Lab. From there, we will select the standard python3 environment
(conda_python3) to start our first .ipynb notebook (“Jupyter notebook”).
You can name your Jupyter notebook something along the lines of,
Interacting-with-S3.ipynb.
We can use the standard conda_python3 environment since we aren’t doing any training/tuning just yet.
Set up AWS environment
To begin each SageMaker notebook, it’s important to set up an AWS environment that will allow seamless access to the necessary cloud resources. Here’s what we’ll do to get started:
Define the Role: We’ll use
get_execution_role()to retrieve the IAM role associated with the SageMaker instance. This role specifies the permissions needed for interacting with AWS services like S3, which allows SageMaker to securely read from and write to storage buckets.Initialize the SageMaker Session: Next, we’ll create a
sagemaker.Session()object, which will help manage and track the resources and operations we use in SageMaker, such as training jobs and model artifacts. The session acts as a bridge between the SageMaker SDK commands in our notebook and AWS services.Set Up an S3 Client: Using
boto3, we’ll initialize an S3 client for accessing S3 buckets directly. This client enables us to handle data storage, retrieve datasets, and manage files in S3, which will be essential as we work through various machine learning tasks.
Starting with these initializations prepares our notebook environment to efficiently interact with AWS resources for model development, data management, and deployment.
Reading data from S3
You can either read data from S3 into memory or download a copy of your S3 data into your notebook’s instance. While loading into memory can save on storage resources, it can be convenient at times to have a local copy. We’ll show you both strategies in this upcoming section. Here’s a more detailed look at the pros and cons of each strategy:
-
Reading data directly from S3 into memory:
-
Pros:
- Storage efficiency: By keeping data in memory, you avoid taking up local storage on your notebook instance, which can be particularly beneficial for larger datasets or instances with limited storage.
- Simple data management: Accessing data directly from S3 avoids the need to manage or clean up local copies after processing.
-
Cons:
- Performance for frequent reads: Accessing S3 data repeatedly can introduce latency and slow down workflows, as each read requires a network request. This approach works best if you only need to load data once or infrequently.
- Potential cost for high-frequency access: Multiple GET requests to S3 can accumulate charges over time, especially if your workflow requires repeated access to the same data.
-
Pros:
-
Downloading a copy of data from S3 to local
storage:
-
Pros:
- Better performance for intensive workflows: If you need to access the dataset multiple times during processing, working from a local copy avoids repeated network requests, making operations faster and more efficient.
- Offline access: Once downloaded, you can access the data without a persistent internet connection, which can be helpful for handling larger data transformations.
-
Cons:
- Storage costs: Local storage on the instance may come with additional costs or limitations, especially if your instance type has constrained storage capacity.
- Data management overhead: You’ll need to manage local data copies and ensure that they are properly cleaned up to free resources once processing is complete.
-
Pros:
Choosing between the two strategies
If your workflow requires only a single read of the dataset for processing, reading directly into memory can be a quick and resource-efficient solution. However, for cases where you’ll perform extensive or iterative processing, downloading a local copy of the data will typically be more performant and may incur fewer request-related costs.
1A. Read data from S3 into memory
Our data is stored on an S3 bucket called ‘titanic-dataset-test’. We can use the following code to read data directly from S3 into memory in the Jupyter notebook environment, without actually downloading a copy of train.csv as a local file.
PYTHON
import pandas as pd
# Define the S3 bucket and object key
bucket_name = 'myawesometeam-titanic' # replace with your S3 bucket name
# Read the train data from S3
key = 'titanic_train.csv' # replace with your object key
response = s3.get_object(Bucket=bucket_name, Key=key)
train_data = pd.read_csv(response['Body'])
# Read the test data from S3
key = 'titanic_test.csv' # replace with your object key
response = s3.get_object(Bucket=bucket_name, Key=key)
test_data = pd.read_csv(response['Body'])
# check shape
print(train_data.shape)
print(test_data.shape)
# Inspect the first few rows of the DataFrame
train_data.head()1B. Download copy into notebook environment
Download data from S3 to notebook environment. You may need to hit refresh on the file explorer panel to the left to see this file. If you get any permission issues…
- check that you have selected the appropriate policy for this notebook
- check that your bucket has the appropriate policy permissions
2. Check current size and storage costs of bucket
It’s a good idea to periodically check how much storage you have used in your bucket. You can do this from a Jupyter notebook in SageMaker by using the Boto3 library, which is the AWS SDK for Python. This will allow you to calculate the total size of objects within a specified bucket. Here’s how you can do it…
Step 1: Set up the S3 Client and Calculate Bucket Size
The code below will calculate your bucket size for you. Here is a breakdown of the important pieces in the next code section:
- Paginator: Since S3 buckets can contain many objects, we use a paginator to handle large listings.
-
Size calculation: We sum the
Sizeattribute of each object in the bucket. -
Unit conversion: The size is given in bytes, so
dividing by
1024 ** 2converts it to megabytes (MB).
Note: If your bucket has very large objects or you want to check specific folders within a bucket, you may want to refine this code to only fetch certain objects or folders.
PYTHON
# Initialize the total size counter
total_size_bytes = 0
# List and sum the size of all objects in the bucket
paginator = s3.get_paginator('list_objects_v2')
for page in paginator.paginate(Bucket=bucket_name):
for obj in page.get('Contents', []):
total_size_bytes += obj['Size']
# Convert the total size to gigabytes for cost estimation
total_size_gb = total_size_bytes / (1024 ** 3)
# print(f"Total size of bucket '{bucket_name}': {total_size_gb:.2f} GB") # can uncomment this if you want GB reported
# Convert the total size to megabytes for readability
total_size_mb = total_size_bytes / (1024 ** 2)
print(f"Total size of bucket '{bucket_name}': {total_size_mb:.2f} MB")Using helper functions from lesson repo
We have added code to calculate bucket size to a helper function
called get_s3_bucket_size(bucket_name) for your
convenience. There are also some other helper functions in that repo to
assist you with common AWS/SageMaker workflows. We’ll show you how to
clone this code into your notebook environment.
Note: Make sure you have already forked the lesson repo as described on the setup page. Replace “username” below with your GitHub username.
Directory setup
Let’s make sure we’re starting in the root directory of this instance, so that we all have our AWS_helpers.py file located in the same path (/test_AWS/scripts/AWS_helpers.py)
To clone the repo to our Jupyter notebook, use the following code.
PYTHON
!git clone https://github.com/username/AWS_helpers.git # downloads AWS_helpers folder/repo (refresh file explorer to see)Our AWS_helpers.py file can be found in
AWS_helpers/helpers.py. With this file downloaded, you can
call this function via…
3: Check storage costs of bucket
To estimate the storage cost of your Amazon S3 bucket directly from a Jupyter notebook in SageMaker, you can use the following approach. This method calculates the total size of the bucket and estimates the monthly storage cost based on AWS S3 pricing.
Note: AWS S3 pricing varies by region and storage class. The example below uses the S3 Standard storage class pricing for the US East (N. Virginia) region as of November 1, 2024. Please verify the current pricing for your specific region and storage class on the AWS S3 Pricing page.
PYTHON
# AWS S3 Standard Storage pricing for US East (N. Virginia) region
# Pricing tiers as of November 1, 2024
first_50_tb_price_per_gb = 0.023 # per GB for the first 50 TB
next_450_tb_price_per_gb = 0.022 # per GB for the next 450 TB
over_500_tb_price_per_gb = 0.021 # per GB for storage over 500 TB
# Calculate the cost based on the size
if total_size_gb <= 50 * 1024:
# Total size is within the first 50 TB
cost = total_size_gb * first_50_tb_price_per_gb
elif total_size_gb <= 500 * 1024:
# Total size is within the next 450 TB
cost = (50 * 1024 * first_50_tb_price_per_gb) + \
((total_size_gb - 50 * 1024) * next_450_tb_price_per_gb)
else:
# Total size is over 500 TB
cost = (50 * 1024 * first_50_tb_price_per_gb) + \
(450 * 1024 * next_450_tb_price_per_gb) + \
((total_size_gb - 500 * 1024) * over_500_tb_price_per_gb)
print(f"Estimated monthly storage cost: ${cost:.4f}")
print(f"Estimated annual storage cost: ${cost*12:.4f}")For your convenience, we have also added this code to a helper function.
PYTHON
monthly_cost, storage_size_gb = helpers.calculate_s3_storage_cost(bucket_name)
print(f"Estimated monthly cost ({storage_size_gb:.4f} GB): ${monthly_cost:.5f}")
print(f"Estimated annual cost ({storage_size_gb:.4f} GB): ${monthly_cost*12:.5f}")Important Considerations:
-
Pricing Tiers: AWS S3 pricing is tiered. The first
50 TB per month is priced at
$0.023 per GB, the next 450 TB at$0.022 per GB, and storage over 500 TB at$0.021 per GB. Ensure you apply the correct pricing tier based on your total storage size. - Region and Storage Class: Pricing varies by AWS region and storage class. The example above uses the S3 Standard storage class pricing for the US East (N. Virginia) region. Adjust the pricing variables if your bucket is in a different region or uses a different storage class.
- Additional Costs: This estimation covers storage costs only. AWS S3 may have additional charges for requests, data retrievals, and data transfers. For a comprehensive cost analysis, consider these factors as well.
For detailed and up-to-date information on AWS S3 pricing, please refer to the AWS S3 Pricing page.
4. Pushing new files from notebook environment to bucket
As your analysis generates new files, you can upload to your bucket
as demonstrated below. For this demo, you can create a blank
results.txt file to upload to your bucket. To do so, go to
File -> New -> Text
file, and save it out as results.txt.
PYTHON
# Define the S3 bucket name and the file paths
train_file_path = "results.txt" # assuming your file is in root directory of jupyter notebook (check file explorer tab)
# Upload the training file to a new folder called "results". You can also just place it in the bucket's root directory if you prefer (remove results/ in code below).
s3.upload_file(train_file_path, bucket_name, "results/results.txt")
print("Files uploaded successfully.")After uploading, we can view the objects/files available on our bucket using…
PYTHON
# List and print all objects in the bucket
response = s3.list_objects_v2(Bucket=bucket_name)
# Check if there are objects in the bucket
if 'Contents' in response:
for obj in response['Contents']:
print(obj['Key']) # Print the object's key (its path in the bucket)
else:
print("The bucket is empty or does not exist.")Alternatively, we can substitute this for a helper function call as well.
[‘results/results.txt’, ‘titanic_test.csv’, ‘titanic_train.csv’]
Key Points
- Load data from S3 into memory for efficient storage and processing.
- Periodically check storage usage and costs to manage S3 budgets.
- Use SageMaker to upload analysis results and maintain an organized workflow.
Content from Using a GitHub Personal Access Token (PAT) to Push/Pull from a SageMaker Notebook
Last updated on 2024-11-06 | Edit this page
Overview
Questions
- How can I securely push/pull code to and from GitHub within a SageMaker notebook?
- What steps are necessary to set up a GitHub PAT for authentication in SageMaker?
- How can I convert notebooks to
.pyfiles and ignore.ipynbfiles in version control?
Objectives
- Configure Git in a SageMaker notebook to use a GitHub Personal Access Token (PAT) for HTTPS-based authentication.
- Securely handle credentials in a notebook environment using
getpass. - Convert
.ipynbfiles to.pyfiles for better version control practices in collaborative projects.
Step 0: Initial setup
In the previous episode, we cloned our fork that we created during the workshop setup. In this episode, we’ll see how to push our code to this fork. Complete these three setup steps before moving foward.
Clone the fork if you haven’t already. See previous episode.
Start a new Jupyter notebook, and name it something along the lines of “Interacting-with-git.ipynb”. We can use the standard conda_python3 environment since we aren’t doing any training/tuning just yet.
Let’s make sure we’re starting at the same directory. Cd to the root directory of this instance before going further.
Step 1: Using a GitHub personal access token (PAT) to push/pull from a SageMaker notebook
When working in SageMaker notebooks, you may often need to push code updates to GitHub repositories. However, SageMaker notebooks are typically launched with temporary instances that don’t persist configurations, including SSH keys, across sessions. This makes HTTPS-based authentication, secured with a GitHub Personal Access Token (PAT), a practical solution. PATs provide flexibility for authentication and enable seamless interaction with both public and private repositories directly from your notebook.
Important Note: Personal access tokens are powerful credentials that grant specific permissions to your GitHub account. To ensure security, only select the minimum necessary permissions and handle the token carefully.
Generate a personal access token (PAT) on GitHub
- Go to Settings > Developer settings > Personal access tokens on GitHub.
- Click Generate new token, select Classic.
- Give your token a descriptive name (e.g., “SageMaker Access Token”) and set an expiration date if desired for added security.
-
Select the minimum permissions needed:
-
For public repositories: Choose only
public_repo. -
For private repositories: Choose
repo(full control of private repositories). - Optional permissions, if needed:
-
repo:status: Access commit status (if checking status checks). -
workflow: Update GitHub Actions workflows (only if working with GitHub Actions).
-
-
For public repositories: Choose only
- Generate the token and copy it (you won’t be able to see it again).
Caution: Treat your PAT like a password. Avoid sharing it or exposing it in your code. Store it securely (e.g., via a password manager like LastPass) and consider rotating it regularly.
Use getpass to prompt for username and PAT
The getpass library allows you to input your GitHub
username and PAT without exposing them in the notebook. This approach
ensures you’re not hardcoding sensitive information.
PYTHON
import getpass
# Prompt for GitHub username and PAT securely
username = input("GitHub Username: ")
token = getpass.getpass("GitHub Personal Access Token (PAT): ")Note: After running, you may want to comment out the above code so that you don’t have to enter in your login every time you run your whole notebook
Step 2: Configure Git settings
In your SageMaker or Jupyter notebook environment, run the following commands to set up your Git user information.
Setting this globally (--global) will ensure the
configuration persists across all repositories in the environment. If
you’re working in a temporary environment, you may need to re-run this
configuration after a restart.
PYTHON
!git config --global user.name "Your name" # This is your GitHub username (or just your name), which will appear in the commit history as the author of the changes.
!git config --global user.email your_email@wisc.edu # This should match the email associated with your GitHub account so that commits are properly linked to your profile.Step 3: Convert json .ipynb files to .py
We’d like to track our notebook files within our AWS_helpers fork.
However, to avoid tracking ipynb files directly, which are formatted as
json, we may want to convert our notebook to .py first (plain text).
Converting notebooks to .py files helps maintain code (and
version-control) readability and minimizes potential issues with
notebook-specific metadata in Git history.
Benefits of converting to .py before Committing
-
Cleaner version control:
.pyfiles have cleaner diffs and are easier to review and merge in Git. - Script compatibility: Python files are more compatible with other environments and can run easily from the command line.
-
Reduced repository size:
.pyfiles are generally lighter than.ipynbfiles since they don’t store outputs or metadata.
Here’s how to convert .ipynb files to .py
in SageMaker without needing to export or download files.
- First, install Jupytext.
- Then, run the following command in a notebook cell to convert both
of our notebooks to
.pyfiles
PYTHON
# Adjust filename(s) if you used something different
!jupytext --to py Interacting-with-S3.ipynbSH
[jupytext] Reading Interacting-with-S3.ipynb in format ipynb
[jupytext] Writing Interacting-with-S3.py- If you have multiple notebooks to convert, you can automate the
conversion process by running this code, which converts all
.ipynbfiles in the current directory to.pyfiles:
PYTHON
import subprocess
import os
# List all .ipynb files in the directory
notebooks = [f for f in os.listdir() if f.endswith('.ipynb')]
# Convert each notebook to .py using jupytext
for notebook in notebooks:
output_file = notebook.replace('.ipynb', '.py')
subprocess.run(["jupytext", "--to", "py", notebook, "--output", output_file])
print(f"Converted {notebook} to {output_file}")For convenience, we have placed this code inside a
convert_files() function in helpers.py.
Once converted, we can move our .py files to the AWS_helpers folder using the file explorer panel in Jupyter Lab.
Step 4. Add and commit .py files
- Check status of repo. Make sure you’re in the repo folder before running the next step.
On branch main Your branch is up to date with ‘origin/main’.
Untracked files: (use “git add
nothing added to commit but untracked files present (use “git add” to track)
- Add and commit changes
PYTHON
!git add . # you may also add files one at a time, for further specificity over the associated commit message
!git commit -m "Updates from Jupyter notebooks" # in general, your commit message should be more specific!- Check status
Step 5. Adding .ipynb to gitigore
Adding .ipynb files to .gitignore is a good
practice if you plan to only commit .py scripts. This will
prevent accidental commits of Jupyter Notebook files across all
subfolders in the repository.
Here’s how to add .ipynb files to
.gitignore to ignore them project-wide:
-
Cd to git repo folder First make sure we’re in the repo folder
Create the
.gitignorefile: This file will be hidden in Jupyter (since it starts with “.”), but you can verify it exists usingls.python !touch .gitignore !ls -a-
Add
.ipynbfiles to.gitignore:You can add this line using a command within your notebook:
PYTHON
with open(".gitignore", "a") as gitignore: gitignore.write("\n# Ignore all Jupyter Notebook files\n*.ipynb\n")View file contents
python !cat .gitignore -
Ignore other common temp files While we’re at it, let’s ignore other common files that can clutter repos, such as cache folders and temporary files.
PYTHON
with open(".gitignore", "a") as gitignore: gitignore.write("\n# Ignore cache and temp files\n__pycache__/\n*.tmp\n*.log\n")View file contents
-
Add and commit the
.gitignorefile:Add and commit the updated
.gitignorefile to ensure it’s applied across the repository.
This setup will:
- Prevent all
.ipynbfiles from being tracked by Git. - Keep your repository cleaner, containing only
.pyscripts for easier version control and reduced repository size.
Step 6. Merging local changes with remote/GitHub
Our local changes have now been committed, and we can begin the process of mergining with the remoate main branch. Before we try to push our changes, it’s good practice to first to a pull. This is critical when working on a collaborate repo with multiple users, so that you don’t miss any updates from other team members.
1. Pull the latest changes from the main branch
There are a few different options for pulling the remote code into your local version. The best pull strategy depends on your workflow and the history structure you want to maintain. Here’s a breakdown to help you decide:
- Merge (pull.rebase false): Combines the remote changes into your
local branch as a merge commit.
- Use if: You’re okay with having merge commits in your history, which indicate where you pulled in remote changes. This is the default and is usually the easiest for team collaborations, especially if conflicts arise.
- Rebase (pull.rebase true): Replays your local changes on top of the
updated main branch, resulting in a linear history.
- Use if: You prefer a clean, linear history without merge commits. Rebase is useful if you like to keep your branch history as if all changes happened sequentially.
- Fast-forward only (pull.ff only): Only pulls if the local branch can
fast-forward to the remote without diverging (no new commits locally).
- Use if: You only want to pull updates if no additional commits have been made locally. This can be helpful to avoid unintended merges when your branch hasn’t diverged.
Recommended for Most Users
If you’re collaborating and want simplicity, merge (pull.rebase false) is often the most practical option. This will ensure you get remote changes with a merge commit that captures the history of integration points. For those who prefer a more streamlined history and are comfortable with Git, rebase (pull.rebase true) can be ideal but may require more careful conflict handling.
PYTHON
!git config pull.rebase false # Combines the remote changes into your local branch as a merge commit.
!git pull origin mainSH
From https://github.com/qualiaMachine/AWS_helpers
* branch main -> FETCH_HEAD
Already up to date.If you get merge conflicts, be sure to resolve those before moving forward (e.g., use git checkout -> add -> commit). You can skip the below code if you don’t have any conflicts.
PYTHON
# Keep your local changes in one conflicting file
# !git checkout --ours Interacting-with-git.py
# Keep remote version for the other conflicting file
# !git checkout --theirs Interacting-with-git.py
# # Stage the files to mark the conflicts as resolved
# !git add Interacting-with-git.py
# # Commit the merge result
# !git commit -m "Resolved merge conflicts by keeping local changes"2. Push changes using PAT creditials
PYTHON
# Push with embedded credentials from getpass (avoids interactive prompt)
github_url = 'github.com/username/AWS_helpers.git' # replace username with your own. THe full address for your fork can be found under Code -> Clone -> HTTPS (remote the https:// before the rest of the address)
!git push https://{username}:{token}@{github_url} mainSH
Enumerating objects: 6, done.
Counting objects: 100% (6/6), done.
Delta compression using up to 2 threads
Compressing objects: 100% (5/5), done.
Writing objects: 100% (5/5), 3.00 KiB | 3.00 MiB/s, done.
Total 5 (delta 0), reused 0 (delta 0), pack-reused 0
To https://github.com/qualiaMachine/AWS_helpers.git
bc47546..6a8bb8b main -> mainAfter pushing, you should navigate back to your fork on GitHub to verify everything worked (e.g., https://github.com/username/AWS_helpers/tree/main)
Step 7: Pulling .py files and converting back to notebook format
Let’s assume you’ve taken a short break from your work, and others on
your team have made updates to your .py files on the remote main branch.
If you’d like to work with notebook files again, you can again use
jupytext to convert your .py files back to
.ipynb.
- First, pull any updates from the remote main branch.
PYTHON
!git config pull.rebase false # Combines the remote changes into your local branch as a merge commit.
!git pull origin main- We can then use jupytext again to convert in the other direction
(.py to .ipynb). This command will create
Interacting-with-S3.ipynbin the current directory, converting the Python script to a Jupyter Notebook format. Jupytext handles the conversion gracefully without expecting the.pyfile to be in JSON format.
PYTHON
# Replace 'your_script.py' with your actual filename
!jupytext --to notebook Interacting-with-S3.py --output Interacting-with-S3.ipynbApplying to all .py files
To convert all of your .py files to notebooks, you can use our helper function as follows
Key Points
- Use a GitHub PAT for HTTPS-based authentication in temporary SageMaker notebook instances.
- Securely enter sensitive information in notebooks using
getpass. - Converting
.ipynbfiles to.pyfiles helps with cleaner version control and easier review of changes. - Adding
.ipynbfiles to.gitignorekeeps your repository organized and reduces storage.
Content from Training Models in SageMaker: Intro
Last updated on 2024-11-07 | Edit this page
Overview
Questions
- What are the differences between local training and SageMaker-managed training?
- How do Estimator classes in SageMaker streamline the training process for various frameworks?
- How does SageMaker handle data and model parallelism, and when should each be considered?
Objectives
- Understand the difference between training locally in a SageMaker notebook and using SageMaker’s managed infrastructure.
- Learn to configure and use SageMaker’s Estimator classes for different frameworks (e.g., XGBoost, PyTorch, SKLearn).
- Understand data and model parallelism options in SageMaker, including when to use each for efficient training.
- Compare performance, cost, and setup between custom scripts and built-in images in SageMaker.
- Conduct training with data stored in S3 and monitor training job status using the SageMaker console.
Initial setup
1. Open a new .ipynb notebook
Open a fresh .ipynb notebook (“Jupyter notebook”), and select the
conda_pytorch_p310 environment. This will save us the trouble of having
to install pytorch in this notebook. You can name your Jupyter notebook
something along the lines of, Training-models.ipynb.
2. CD to instance home directory
So we all can reference the helper functions using the same path, CD to…
3. Initialize SageMaker environment
This code initializes the AWS SageMaker environment by defining the SageMaker role, session, and S3 client. It also specifies the S3 bucket and key for accessing the Titanic training dataset stored in an S3 bucket.
Boto3 API
Boto3 is the official AWS SDK for Python, allowing developers to interact programmatically with AWS services like S3, EC2, and Lambda. It provides both high-level and low-level APIs, making it easy to manage AWS resources and automate tasks. With built-in support for paginators, waiters, and session management, Boto3 simplifies working with AWS credentials, regions, and IAM permissions. It’s ideal for automating cloud operations and integrating AWS services into Python applications.
PYTHON
import boto3
import pandas as pd
import sagemaker
from sagemaker import get_execution_role
# Initialize the SageMaker role (will reflect notebook instance's policy)
role = sagemaker.get_execution_role()
print(f'role = {role}')
# Create a SageMaker session to manage interactions with Amazon SageMaker, such as training jobs, model deployments, and data input/output.
session = sagemaker.Session()
# Initialize an S3 client to interact with Amazon S3, allowing operations like uploading, downloading, and managing objects and buckets.
s3 = boto3.client('s3')
# Define the S3 bucket that we will load from
bucket_name = 'myawesometeam-titanic' # replace with your S3 bucket name
# Define train/test filenames
train_filename = 'titanic_train.csv'
test_filename = 'titanic_test.csv'3. Download copy into notebook environment
It can be convenient to have a “local” copy (i.e., one that you store in your notebook’s instance). Run the next code chunk to download data from S3 to notebook environment. You may need to hit refresh on the file explorer panel to the left to see this file. If you get any permission issues…
- check that you have selected the appropriate policy for this notebook
- check that your bucket has the appropriate policy permissions
PYTHON
# Define the S3 bucket and file location
file_key = f"{train_filename}" # Path to your file in the S3 bucket
local_file_path = f"./{train_filename}" # Local path to save the file
# Download the file using the s3 client variable we initialized earlier
s3.download_file(bucket_name, file_key, local_file_path)
print("File downloaded:", local_file_path)We can do the same for the test set.
PYTHON
# Define the S3 bucket and file location
file_key = f"{test_filename}" # Path to your file in the S3 bucket. W
local_file_path = f"./{test_filename}" # Local path to save the file
# Initialize the S3 client and download the file
s3.download_file(bucket_name, file_key, local_file_path)
print("File downloaded:", local_file_path)Testing train.py on this notebook’s instance
In this next section, we will learn how to take a model training script, and deploy it to more powerful instances (or many instances). This is helpful for machine learning jobs that require extra power, GPUs, or benefit from parallelization. Before we try exploiting this extra power, it is essential that we test our code thoroughly. We don’t want to waste unnecessary compute cycles and resources on jobs that produce bugs instead of insights. If you need to, you can use a subset of your data to run quicker tests. You can also select a slightly better instance resource if your current instance insn’t meeting your needs. See the Instances for ML spreadsheet for guidance.
Logging runtime & instance info
To compare our local runtime with future experiments, we’ll need to know what instance was used, as this will greatly impact runtime in many cases. We can extract the instance name for this notebook using…
PYTHON
# Replace with your notebook instance name.
# This does NOT refer to specific ipynb files, but to the SageMaker notebook instance.
notebook_instance_name = 'MyAwesomeTeam-ChrisEndemann-Titanic-Train-Tune-Xgboost-NN'
# Initialize SageMaker client
sagemaker_client = boto3.client('sagemaker')
# Describe the notebook instance
response = sagemaker_client.describe_notebook_instance(NotebookInstanceName=notebook_instance_name)
# Display the status and instance type
print(f"Notebook Instance '{notebook_instance_name}' status: {response['NotebookInstanceStatus']}")
local_instance = response['InstanceType']
print(f"Instance Type: {local_instance}")Helper: get_notebook_instance_info()
You can also use the get_notebook_instance_info()
function found in AWS_helpers.py to retrieve this info for
your own project.
PYTHON
import AWS_helpers.helpers as helpers
helpers.get_notebook_instance_info(notebook_instance_name)Test train.py on this notebook’s instance (or when possible, on your own machine) before doing anything more complicated (e.g., hyperparameter tuning on multiple instances)
SH
Collecting xgboost
Downloading xgboost-2.1.2-py3-none-manylinux2014_x86_64.whl.metadata (2.0 kB)
Requirement already satisfied: numpy in /home/ec2-user/anaconda3/envs/pytorch_p310/lib/python3.10/site-packages (from xgboost) (1.26.4)
Requirement already satisfied: scipy in /home/ec2-user/anaconda3/envs/pytorch_p310/lib/python3.10/site-packages (from xgboost) (1.14.1)
Downloading xgboost-2.1.2-py3-none-manylinux2014_x86_64.whl (4.5 MB)
━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 4.5/4.5 MB 82.5 MB/s eta 0:00:00
Installing collected packages: xgboost
Successfully installed xgboost-2.1.2Local test
PYTHON
import time as t # we'll use the time package to measure runtime
start_time = t.time()
# Define your parameters. These python vars wil be passed as input args to our train_xgboost.py script using %run
max_depth = 3 # Sets the maximum depth of each tree in the model to 3. Limiting tree depth helps control model complexity and can reduce overfitting, especially on small datasets.
eta = 0.1 # Sets the learning rate to 0.1, which scales the contribution of each tree to the final model. A smaller learning rate often requires more rounds to converge but can lead to better performance.
subsample = 0.8 # Specifies that 80% of the training data will be randomly sampled to build each tree. Subsampling can help with model robustness by preventing overfitting and increasing variance.
colsample_bytree = 0.8 # Specifies that 80% of the features will be randomly sampled for each tree, enhancing the model's ability to generalize by reducing feature reliance.
num_round = 100 # Sets the number of boosting rounds (trees) to 100. More rounds typically allow for a more refined model, but too many rounds can lead to overfitting.
train_file = 'titanic_train.csv' # Points to the location of the training data
# Use f-strings to format the command with your variables
%run AWS_helpers/train_xgboost.py --max_depth {max_depth} --eta {eta} --subsample {subsample} --colsample_bytree {colsample_bytree} --num_round {num_round} --train {train_file}
# Measure and print the time taken
print(f"Total local runtime: {t.time() - start_time:.2f} seconds, instance_type = {local_instance}")SH
Train size: (569, 8)
Val size: (143, 8)
Training time: 0.06 seconds
Model saved to ./xgboost-model
Total local runtime: 1.01 seconds, instance_type = ml.t3.medium
/home/ec2-user/anaconda3/envs/pytorch_p310/lib/python3.10/site-packages/xgboost/core.py:265: FutureWarning: Your system has an old version of glibc (< 2.28). We will stop supporting Linux distros with glibc older than 2.28 after **May 31, 2025**. Please upgrade to a recent Linux distro (with glibc 2.28+) to use future versions of XGBoost.
Note: You have installed the 'manylinux2014' variant of XGBoost. Certain features such as GPU algorithms or federated learning are not available. To use these features, please upgrade to a recent Linux distro with glibc 2.28+, and install the 'manylinux_2_28' variant.
warnings.warn(Training on this relatively small dataset should take less than a minute, but as we scale up with larger datasets and more complex models in SageMaker, tracking both training time and total runtime becomes essential for efficient debugging and resource management.
Note: Our code above includes print statements to monitor dataset size, training time, and total runtime, which provides insights into resource usage for model development. We recommend incorporating similar logging to track not only training time but also total runtime, which includes additional steps like data loading, evaluation, and saving results. Tracking both can help you pinpoint bottlenecks and optimize your workflow as projects grow in size and complexity, especially when scaling with SageMaker’s distributed resources.
Quick evaluation on test set
This next section isn’t SageMaker specific, so we’ll cover it quickly. Here’s how you would apply the outputted model to your test set using your local notebook instance.
PYTHON
import xgboost as xgb
import pandas as pd
import numpy as np
from sklearn.metrics import accuracy_score
import joblib
from AWS_helpers.train_xgboost import preprocess_data
# Load the test data
test_data = pd.read_csv('./titanic_test.csv')
# Preprocess the test data using the imported preprocess_data function
X_test, y_test = preprocess_data(test_data)
# Convert the test features to DMatrix for XGBoost
dtest = xgb.DMatrix(X_test)
# Load the trained model from the saved file
model = joblib.load('./xgboost-model')
# Make predictions on the test set
preds = model.predict(dtest)
predictions = np.round(preds) # Round predictions to 0 or 1 for binary classification
# Calculate and print the accuracy of the model on the test data
accuracy = accuracy_score(y_test, predictions)
print(f"Test Set Accuracy: {accuracy:.4f}")Training via SageMaker (using notebook as controller) - custom train.py script
Unlike “local” training (using this notebook), this next approach leverages SageMaker’s managed infrastructure to handle resources, parallelism, and scalability. By specifying instance parameters, such as instance_count and instance_type, you can control the resources allocated for training.
Which instance to start with?
In this example, we start with one ml.m5.large instance, which is suitable for small- to medium-sized datasets and simpler models. Using a single instance is often cost-effective and sufficient for initial testing, allowing for straightforward scaling up to more powerful instance types or multiple instances if training takes too long. See here for further guidance on selecting an appropriate instance for your data/model: EC2 Instances for ML
Overview of Estimator classes in SageMaker
To launch this training “job”, we’ll use the XGBoost “Estimator. In
SageMaker, Estimator classes streamline the configuration and training
of models on managed instances. Each Estimator can work with custom
scripts and be enhanced with additional dependencies by specifying a
requirements.txt file, which is automatically installed at
the start of training. Here’s a breakdown of some commonly used
Estimator classes in SageMaker:
1. Estimator (Base Class)
- Purpose: General-purpose for custom Docker containers or defining an image URI directly.
-
Configuration: Requires specifying an
image_uriand custom entry points. -
Dependencies: You can use
requirements.txtto install Python packages or configure a custom Docker container with pre-baked dependencies. - Ideal Use Cases: Custom algorithms or models that need tailored environments not covered by built-in containers.
2. XGBoost Estimator
- Purpose: Provides an optimized container specifically for XGBoost models.
-
Configuration:
-
entry_point: Path to a custom script, useful for additional preprocessing or unique training workflows. -
framework_version: Select XGBoost version, e.g.,"1.5-1". -
dependencies: Specify additional packages throughrequirements.txtto enhance preprocessing capabilities or incorporate auxiliary libraries.
-
- Ideal Use Cases: Tabular data modeling using gradient-boosted trees; cases requiring custom preprocessing or tuning logic.
3. PyTorch Estimator
- Purpose: Configures training jobs with PyTorch for deep learning tasks.
-
Configuration:
-
entry_point: Training script with model architecture and training loop. -
instance_type: e.g.,ml.p3.2xlargefor GPU acceleration. -
framework_versionandpy_version: Define specific versions. -
dependencies: Install any required packages viarequirements.txtto support advanced data processing, data augmentation, or custom layer implementations.
-
- Ideal Use Cases: Deep learning models, particularly complex networks requiring GPUs and custom layers.
4. SKLearn Estimator
- Purpose: Supports scikit-learn workflows for data preprocessing and classical machine learning.
-
Configuration:
-
entry_point: Python script to handle feature engineering, preprocessing, or training. -
framework_version: Version of scikit-learn, e.g.,"1.0-1". -
dependencies: Userequirements.txtto install any additional Python packages required by the training script.
-
- Ideal Use Cases: Classical ML workflows, extensive preprocessing, or cases where additional libraries (e.g., pandas, numpy) are essential.
5. TensorFlow Estimator
- Purpose: Designed for training and deploying TensorFlow models.
-
Configuration:
-
entry_point: Script for model definition and training process. -
instance_type: Select based on dataset size and computational needs. -
dependencies: Additional dependencies can be listed inrequirements.txtto install TensorFlow add-ons, custom layers, or preprocessing libraries.
-
- Ideal Use Cases: NLP, computer vision, and transfer learning applications in TensorFlow.
Configuring custom environments with
requirements.txt
For all these Estimators, adding a requirements.txt file
under dependencies ensures that additional packages are
installed before training begins. This approach allows the use of
specific libraries that may be critical for custom preprocessing,
feature engineering, or model modifications. Here’s how to include
it:
PYTHON
sklearn_estimator = SKLearn(
entry_point="train_script.py",
role=role,
instance_count=1,
instance_type="ml.m5.large",
output_path="s3://your-bucket/output",
framework_version="1.0-1",
dependencies=['requirements.txt'], # Adding custom dependencies
hyperparameters={
"max_depth": 5,
"eta": 0.1,
"subsample": 0.8,
"num_round": 100
}
)This setup simplifies training, allowing you to maintain custom environments directly within SageMaker’s managed containers, without needing to build and manage your own Docker images. The AWS SageMaker Documentation provides lists of pre-built container images for each framework and their standard libraries, including details on pre-installed packages.
Deploying to other instances
For this deployment, we configure the “XGBoost” estimator with a custom training script, train_xgboost.py, and define hyperparameters directly within the SageMaker setup. Here’s the full code, with some additional explanation following the code.
PYTHON
from sagemaker.inputs import TrainingInput
from sagemaker.xgboost.estimator import XGBoost
# Define instance type/count we'll use for training
instance_type="ml.m5.large"
instance_count=1 # always start with 1. Rarely is parallelized training justified with data < 50 GB. More on this later.
# Define S3 paths for input and output
train_s3_path = f's3://{bucket_name}/{train_filename}'
# we'll store all results in a subfolder called xgboost on our bucket. This folder will automatically be created if it doesn't exist already.
output_folder = 'xgboost'
output_path = f's3://{bucket_name}/{output_folder}/'
# Set up the SageMaker XGBoost Estimator with custom script
xgboost_estimator = XGBoost(
entry_point='train_xgboost.py', # Custom script path
source_dir='AWS_helpers', # Directory where your script is located
role=role,
instance_count=instance_count,
instance_type=instance_type,
output_path=output_path,
sagemaker_session=session,
framework_version="1.5-1", # Use latest supported version for better compatibility
hyperparameters={
'train': train_file,
'max_depth': max_depth,
'eta': eta,
'subsample': subsample,
'colsample_bytree': colsample_bytree,
'num_round': num_round
}
)
# Define input data
train_input = TrainingInput(train_s3_path, content_type='csv')
# Measure and start training time
start = t.time()
xgboost_estimator.fit({'train': train_input})
end = t.time()
print(f"Runtime for training on SageMaker: {end - start:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")Hyperparameters
The hyperparameters section in this code defines the
input arguments of train_XGBoost.py. The first is the name of the
training input file, and the others are hyperparameters for the XGBoost
model, such as max_depth, eta,
subsample, colsample_bytree, and
num_round.
TrainingInput
Additionally, we define a TrainingInput object containing the
training data’s S3 path, to pass to
.fit({'train': train_input}). SageMaker uses
TrainingInput to download your dataset from S3 to a
temporary location on the training instance. This location is mounted
and managed by SageMaker and can be accessed by the training job if/when
needed.
Model results
With this code, the training results and model artifacts are saved in
a subfolder called xgboost in your specified S3 bucket.
This folder (s3://{bucket_name}/xgboost/) will be
automatically created if it doesn’t already exist, and will contain:
-
Model “artifacts”: The trained model file (often a
.tar.gzfile) that SageMaker saves in theoutput_path. -
Logs and metrics: Any metrics and logs related to
the training job, stored in the same
xgboostfolder.
This setup allows for convenient access to both the trained model and related output for later evaluation or deployment.
Extracting trained model from S3 for final evaluation
To evaluate the model on a test set after training, we’ll go through these steps:
- Download the trained model from S3.
- Load and preprocess the test dataset.
- Evaluate the model on the test data.
Here’s how you can implement this in your SageMaker notebook. The following code will:
- Download the
model.tar.gzfile containing the trained model from S3. - Load the
test.csvdata from S3 and preprocess it as needed. - Use the XGBoost model to make predictions on the test set and then compute accuracy or other metrics on the results.
If additional metrics or custom evaluation steps are needed, you can add them in place of or alongside accuracy.
PYTHON
# Model results are saved in auto-generated folders. Use xgboost_estimator.latest_training_job.name to get the folder name
model_s3_path = f'{output_folder}/{xgboost_estimator.latest_training_job.name}/output/model.tar.gz'
print(model_s3_path)
local_model_path = 'model.tar.gz'
# Download the trained model from S3
s3.download_file(bucket_name, model_s3_path, local_model_path)
# Extract the model file
import tarfile
with tarfile.open(local_model_path) as tar:
tar.extractall()PYTHON
import xgboost as xgb
import pandas as pd
import numpy as np
from sklearn.metrics import accuracy_score
import joblib
from AWS_helpers.train_xgboost import preprocess_data
# Load the test data
test_data = pd.read_csv('./titanic_test.csv')
# Preprocess the test data using the imported preprocess_data function
X_test, y_test = preprocess_data(test_data)
# Convert the test features to DMatrix for XGBoost
dtest = xgb.DMatrix(X_test)
# Load the trained model from the saved file
model = joblib.load('./xgboost-model')
# Make predictions on the test set
preds = model.predict(dtest)
predictions = np.round(preds) # Round predictions to 0 or 1 for binary classification
# Calculate and print the accuracy of the model on the test data
accuracy = accuracy_score(y_test, predictions)
print(f"Test Set Accuracy: {accuracy:.4f}")Now that we’ve covered training using a custom script with the
XGBoost estimator, let’s examine the built-in image-based
approach. Using SageMaker’s pre-configured XGBoost image streamlines the
setup by eliminating the need to manage custom scripts for common
workflows, and it can also provide optimization advantages. Below, we’ll
discuss both the code and pros and cons of the image-based setup
compared to the custom script approach.
Training with SageMaker’s Built-in XGBoost Image
With the SageMaker-provided XGBoost container, you can bypass custom script configuration if your workflow aligns with standard XGBoost training. This setup is particularly useful when you need quick, default configurations without custom preprocessing or additional libraries.
Comparison: Custom Script vs. Built-in Image
| Feature | Custom Script (XGBoost with
entry_point) |
Built-in XGBoost Image |
|---|---|---|
| Flexibility | Allows for custom preprocessing, data transformation, or advanced
parameterization. Requires a Python script and custom dependencies can
be added through requirements.txt. |
Limited to XGBoost’s built-in functionality, no custom preprocessing steps without additional customization. |
| Simplicity | Requires setting up a script with entry_point and
managing dependencies. Ideal for specific needs but requires
configuration. |
Streamlined for fast deployment without custom code. Simple setup and no need for custom scripts. |
| Performance | Similar performance, though potential for overhead with additional preprocessing. | Optimized for typical XGBoost tasks with faster startup. May offer marginally faster time-to-first-train. |
| Use Cases | Ideal for complex workflows requiring unique preprocessing steps or when adding specific libraries or functionalities. | Best for quick experiments, standard workflows, or initial testing on datasets without complex preprocessing. |
When to use each approach: - Custom script: Recommended if you need to implement custom data preprocessing, advanced feature engineering, or specific workflow steps that require more control over training. - Built-in image: Ideal when running standard XGBoost training, especially for quick experiments or production deployments where default configurations suffice.
Both methods offer powerful and flexible approaches to model training on SageMaker, allowing you to select the approach best suited to your needs. Below is an example of training using the built-in XGBoost Image.
Setting up the data path
In this approach, using TrainingInput directly with
SageMaker’s built-in XGBoost container contrasts with our previous
method, where we specified a custom script with argument inputs
(specified in hyperparameters) for data paths and settings. Here, we use
hyperparameters only to specify the model’s hyperparameters.
PYTHON
from sagemaker.estimator import Estimator # when using images, we use the general Estimator class
# Define instance type/count we'll use for training
instance_type="ml.m5.large"
instance_count=1 # always start with 1. Rarely is parallelized training justified with data < 50 GB. More on this later.
# Use Estimator directly for built-in container without specifying entry_point
xgboost_estimator_builtin = Estimator(
image_uri=sagemaker.image_uris.retrieve("xgboost", session.boto_region_name, version="1.5-1"),
role=role,
instance_count=instance_count,
instance_type=instance_type,
output_path=output_path,
sagemaker_session=session,
hyperparameters={
'max_depth': max_depth,
'eta': eta,
'subsample': subsample,
'colsample_bytree': colsample_bytree,
'num_round': num_round
}
)
# Define input data
train_input = TrainingInput(train_s3_path, content_type="csv")
# Measure and start training time
start = t.time()
xgboost_estimator_builtin.fit({'train': train_input})
end = t.time()
print(f"Runtime for training on SageMaker: {end - start:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")Monitoring training
To view and monitor your SageMaker training job, follow these steps in the AWS Management Console. Since training jobs may be visible to multiple users in your account, it’s essential to confirm that you’re interacting with your own job before making any changes.
-
Navigate to the SageMaker Console
- Go to the AWS Management Console and open the SageMaker service (can search for it)
-
View training jobs
- In the left-hand navigation menu, select Training jobs. You’ll see a list of recent training jobs, which may include jobs from other users in the account.
-
Verify your training Job
- Identify your job by looking for the specific name format (e.g.,
sagemaker-xgboost-YYYY-MM-DD-HH-MM-SS-XXX) generated when you launched the job. Click on its name to access detailed information. Cross-check the job details, such as the Instance Type and Input data configuration, with the parameters you set in your script.
- Identify your job by looking for the specific name format (e.g.,
-
Monitor the job status
- Once you’ve verified the correct job, click on its name to access
detailed information:
-
Status: Confirms whether the job is
InProgress,Completed, orFailed. - Logs: Review CloudWatch Logs and Job Metrics for real-time updates.
- Output Data: Shows the S3 location with the trained model artifacts.
-
Status: Confirms whether the job is
- Once you’ve verified the correct job, click on its name to access
detailed information:
-
Stopping a training job
- Before stopping a job, ensure you’ve selected the correct one by verifying job details as outlined above.
- If you’re certain it’s your job, go to Training jobs in the SageMaker Console, select the job, and choose Stop from the Actions menu. Confirm your selection, as this action will halt the job and release any associated resources.
- Important: Avoid stopping jobs you don’t own, as this could disrupt other users’ work and may have unintended consequences.
Following these steps helps ensure you only interact with and modify jobs you own, reducing the risk of impacting other users’ training processes.
When training takes too long
When training time becomes excessive, two main options can improve efficiency in SageMaker.
- Option 1: Upgrading to a more powerful instance
- Option 2: Using multiple instances for distributed training.
Generally, Option 1 is the preferred approach and should be explored first.
Option 1: Upgrade to a more powerful instance (preferred starting point)
Upgrading to a more capable instance, particularly one with GPU capabilities (e.g., for deep learning), is often the simplest and most cost-effective way to speed up training. Here’s a breakdown of instances to consider. Check the Instances for ML spreadsheet for guidance on selecting a better instance.
When to use a single instance upgrade
Upgrading a single instance works well if:
- Dataset size: The dataset is small to moderate (e.g., <10 GB), fitting comfortably within the memory of a larger instance.
- Model complexity: The model is not so large that it requires distribution across multiple devices.
- Training time: Expected training time is within a few hours, but could benefit from additional power.
Upgrading a single instance is typically the most efficient option in terms of both cost and setup complexity. It avoids the communication overhead associated with multi-instance setups (discussed below) and is well-suited for most small to medium-sized datasets.
Option 2: Use multiple instances for distributed training
If upgrading a single instance doesn’t sufficiently reduce training time, distributed training across multiple instances may be a viable alternative, particularly for larger datasets and complex models. SageMaker supports two primary distributed training techniques: data parallelism and model parallelism.
Understanding data parallelism vs. model parallelism
Data parallelism: This approach splits the dataset across multiple instances, allowing each instance to process a subset of the data independently. After each batch, gradients are synchronized across instances to ensure consistent updates to the model. Data parallelism is effective when the model itself fits within an instance’s memory, but the data size or desired training speed requires faster processing through multiple instances.
Model parallelism: Model parallelism divides the model itself across multiple instances, making it ideal for very large models (e.g., deep learning models in NLP or image processing) that cannot fit in memory on a single instance. Each instance processes a segment of the model, and results are combined during training. This approach is suitable for memory-intensive models that exceed the capacity of a single instance.
How SageMaker chooses between data and model parallelism
In SageMaker, the choice between data and model parallelism is not entirely automatic. Here’s how it typically works:
Data parallelism (automatic): When you set
instance_count > 1, SageMaker will automatically apply data parallelism. This splits the dataset across instances, allowing each instance to process a subset independently and synchronize gradients after each batch. Data parallelism works well when the model can fit in the memory of a single instance, but the data size or processing speed needs enhancement with multiple instances.Model parallelism (manual setup): To enable model parallelism, you need to configure it explicitly using the SageMaker Model Parallel Library, suitable for deep learning models in frameworks like PyTorch or TensorFlow. Model parallelism splits the model itself across multiple instances, which is useful for memory-intensive models that exceed the capacity of a single instance. Configuring model parallelism requires setting up a distribution strategy in SageMaker’s Python SDK.
Hybrid parallelism (manual setup): For extremely large datasets and models, SageMaker can support both data and model parallelism together, but this setup requires manual configuration. Hybrid parallelism is beneficial for workloads that are both data- and memory-intensive, where both the model and the data need distributed processing.
When to use distributed training with multiple
instances
Consider multiple instances if:
- Dataset size: The dataset is large (>10 GB) and doesn’t fit comfortably within a single instance’s memory.
- Model complexity: The model is complex, requiring extensive computation that a single instance cannot handle in a reasonable time.
- Expected training time: Training on a single instance takes prohibitively long (e.g., >10 hours), and distributed computing overhead is manageable.
Cost of distributed computing
tl;dr Use 1 instance unless you are finding that you’re waiting hours for the training/tuning to complete.
Let’s break down some key points for deciding between 1 instance vs. multiple instances from a cost perspective:
-
Instance cost per hour:
- SageMaker charges per instance-hour. Running multiple instances in parallel can finish training faster, reducing wall-clock time, but the cost per hour will increase with each added instance.
-
Single instance vs. multiple instance wall-clock
time:
- When using a single instance, training will take significantly longer, especially if your data is large. However, the wall-clock time difference between 1 instance and 10 instances may not translate to a direct 10x speedup when using multiple instances due to communication overheads.
- For example, with data-parallel training, instances need to synchronize gradients between batches, which introduces communication costs and may slow down training on larger clusters.
-
Scaling efficiency:
- Parallelizing training does not scale perfectly due to those overheads. Adding instances generally provides diminishing returns on training time reduction.
- For example, doubling instances from 1 to 2 may reduce training time by close to 50%, but going from 8 to 16 instances may only reduce training time by around 20-30%, depending on the model and batch sizes.
-
Typical recommendation:
- For small-to-moderate datasets or cases where training time isn’t a critical factor, a single instance may be more cost-effective, as it avoids parallel processing overheads.
- For large datasets or where training speed is a high priority (e.g., tuning complex deep learning models), using multiple instances can be beneficial despite the cost increase due to time savings.
-
Practical cost estimation:
- Suppose a single instance takes
Thours to train and costs$Cper hour. For a 10-instance setup, the cost would be approximately:-
Single instance:
T * $C -
10 instances (parallel):
(T / k) * (10 * $C), wherekis the speedup factor (<10 due to overhead).
-
Single instance:
- If the speedup is only about 5x instead of 10x due to communication overhead, then the cost difference may be minimal, with a slight edge to a single instance on total cost but at a higher wall-clock time.
- Suppose a single instance takes
In summary: - Start by upgrading to a more powerful instance (Option 1) for datasets up to 10 GB and moderately complex models. A single, more powerful, instance is usually more cost-effective for smaller workloads and where time isn’t critical. Running initial tests with a single instance can also provide a benchmark. You can then experiment with small increases in instance count to find a balance between cost and time savings, particularly considering communication overheads that affect parallel efficiency. - Consider distributed training across multiple instances (Option 2) only when dataset size, model complexity, or training time demand it.
XGBoost’s distributed training mechanism
In the event that option 2 explained above really is better for your use-case (e.g., you have a very large dataset or model that takes a while to train even with high performance instances), the next example will demo setting this up. Before we do, though, we should ask what distributed computing really means for our specific model/setup. XGBoost’s distributed training relies on a data-parallel approach that divides the dataset across multiple instances (or workers), enabling each instance to work on a portion of the data independently. This strategy enhances efficiency, especially for large datasets and computationally intensive tasks.
What about a model parallelism approach? Unlike deep learning models with vast neural network layers, XGBoost’s decision trees are usually small enough to fit in memory on a single instance, even when the dataset is large. Thus, model parallelism is rarely necessary. XGBoost does not inherently support model parallelism out of the box in SageMaker because the model architecture doesn’t typically exceed memory limits, unlike massive language or image models. Although model parallelism can be theoretically applied (e.g., splitting large tree structures across instances), it’s generally not supported natively in SageMaker for XGBoost, as it would require a custom distribution framework to split the model itself.
Here’s how distributed training in XGBoost works, particularly in the SageMaker environment:
Key steps in distributed training with XGBoost
1. Data partitioning
- The dataset is divided among multiple instances. For example, with two instances, each instance may receive half of the dataset.
- In SageMaker, data partitioning across instances is handled automatically via the input channels you specify during training, reducing manual setup.
2. Parallel gradient boosting
- XGBoost performs gradient boosting by constructing trees iteratively based on calculated gradients.
- Each instance calculates gradients (first-order derivatives) and Hessians (second-order derivatives of the loss function) independently on its subset of data.
- This parallel processing allows each instance to determine which features to split and which trees to add to the model based on its data portion.
3. Communication between instances
- After computing gradients and Hessians locally, instances synchronize to share and combine these values.
- Synchronization keeps the model parameters consistent across instances. Only computed gradients are communicated, not the raw dataset, minimizing data transfer overhead.
- The combined gradients guide global model updates, ensuring that the ensemble of trees reflects the entire dataset, despite its division across multiple instances.
4. Final model aggregation
- Once training completes, XGBoost aggregates the trained trees from each instance into a single final model.
- This aggregation enables the final model to perform as though it trained on the entire dataset, even if the dataset couldn’t fit into a single instance’s memory.
SageMaker simplifies these steps by automatically managing the partitioning, synchronization, and aggregation processes during distributed training with XGBoost.
Implementing distributed training with XGBoost in SageMaker
In SageMaker, setting up distributed training for XGBoost can offer significant time savings as dataset sizes and computational requirements increase. Here’s how you can configure it:
-
Select multiple instances: Specify
instance_count > 1in the SageMakerEstimatorto enable distributed training. - Optimize instance type: Choose an instance type suitable for your dataset size and XGBoost requirements
- Monitor for speed improvements: With larger datasets, distributed training can yield time savings by scaling horizontally. However, gains may vary depending on the dataset and computation per instance.
PYTHON
# Define instance type/count we'll use for training
instance_type="ml.m5.large"
instance_count=1 # always start with 1. Rarely is parallelized training justified with data < 50 GB.
# Define the XGBoost estimator for distributed training
xgboost_estimator = Estimator(
image_uri=sagemaker.image_uris.retrieve("xgboost", session.boto_region_name, version="1.5-1"),
role=role,
instance_count=instance_count, # Start with 1 instance for baseline
instance_type=instance_type,
output_path=output_path,
sagemaker_session=session,
)
# Set hyperparameters
xgboost_estimator.set_hyperparameters(
max_depth=5,
eta=0.1,
subsample=0.8,
colsample_bytree=0.8,
num_round=100,
)
# Specify input data from S3
train_input = TrainingInput(train_s3_path, content_type="csv")
# Run with 1 instance
start1 = t.time()
xgboost_estimator.fit({"train": train_input})
end1 = t.time()
# Now run with 2 instances to observe speedup
xgboost_estimator.instance_count = 2
start2 = t.time()
xgboost_estimator.fit({"train": train_input})
end2 = t.time()
print(f"Runtime for training on SageMaker: {end1 - start1:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")
print(f"Runtime for training on SageMaker: {end2 - start2:.2f} seconds, instance_type: {instance_type}, instance_count: {xgboost_estimator.instance_count}")SH
INFO:sagemaker.image_uris:Ignoring unnecessary instance type: None.
INFO:sagemaker:Creating training-job with name: sagemaker-xgboost-2024-11-03-21-16-39-216
2024-11-03 21:16:40 Starting - Starting the training job...
2024-11-03 21:16:55 Starting - Preparing the instances for training...
2024-11-03 21:17:22 Downloading - Downloading input data...
2024-11-03 21:18:07 Downloading - Downloading the training image......
2024-11-03 21:19:13 Training - Training image download completed. Training in progress.
2024-11-03 21:19:13 Uploading - Uploading generated training model[34m/miniconda3/lib/python3.8/site-packages/xgboost/compat.py:36: FutureWarning:
2024-11-03 21:19:32 Completed - Training job completed
INFO:sagemaker:Creating training-job with name: sagemaker-xgboost-2024-11-03-21-19-57-254
Training seconds: 130
Billable seconds: 130
2024-11-03 21:19:58 Starting - Starting the training job...
2024-11-03 21:20:13 Starting - Preparing the instances for training...
2024-11-03 21:20:46 Downloading - Downloading input data......
2024-11-03 21:21:36 Downloading - Downloading the training image...
2024-11-03 21:22:27 Training - Training image download completed. Training in progress..[35m/miniconda3/lib/python3.8/site-packages/xgboost/compat.py:36:
2024-11-03 21:23:01 Uploading - Uploading generated training model
2024-11-03 21:23:01 Completed - Training job completed
Training seconds: 270
Billable seconds: 270
Runtime for training on SageMaker: 198.04 seconds, instance_type: ml.m5.large, instance_count: 1
Runtime for training on SageMaker: 197.66 seconds, instance_type: ml.m5.large, instance_count: 2Why scaling instances might not show speedup here
Small dataset: With only 892 rows, the dataset might be too small to benefit from distributed training. Distributing small datasets often adds overhead (like network communication between instances), which outweighs the parallel processing benefits.
Distributed overhead: Distributed training introduces coordination steps that can add latency. For very short training jobs, this overhead can become a larger portion of the total training time, reducing the benefit of additional instances.
Tree-based models: Tree-based models, like those in XGBoost, don’t benefit from distributed scaling as much as deep learning models when datasets are small. For large datasets, distributed XGBoost can still offer speedups, but this effect is generally less than with neural networks, where parallel gradient updates across multiple instances become efficient.
When multi-instance training helps
Larger datasets: Multi-instance training shines with larger datasets, where splitting the data across instances and processing it in parallel can significantly reduce the training time.
Complex models: For highly complex models with many parameters (like deep learning models or large XGBoost ensembles) and long training times, distributing the training can help speed up the process as each instance contributes to the gradient calculation and optimization steps.
Distributed algorithms: XGBoost has a built-in distributed training capability, but models that perform gradient descent, like deep neural networks, gain more obvious benefits because each instance can compute gradients for a batch of data simultaneously, allowing faster convergence.
For cost optimization
- Single-instance training is typically more cost-effective for small or moderately sized datasets, while multi-instance setups can reduce wall-clock time for larger datasets and complex models, at a higher instance cost.
- For initial testing, start with data parallelism on a single instance, and increase instance count if training time becomes prohibitive, while being mindful of communication overhead and scaling efficiency.
Key Points
- Environment initialization: Setting up a SageMaker session, defining roles, and specifying the S3 bucket are essential for managing data and running jobs in SageMaker.
- Local vs. managed training: Always test your code locally (on a smaller scale) before scaling things up. This avoids wasting resources on buggy code that doesn’t produce reliable results.
- Estimator classes: SageMaker provides framework-specific Estimator classes (e.g., XGBoost, PyTorch, SKLearn) to streamline training setups, each suited to different model types and workflows.
- Custom scripts vs. built-in images: Custom training scripts offer flexibility with preprocessing and custom logic, while built-in images are optimized for rapid deployment and simpler setups.
-
Training data channels: Using
TrainingInputensures SageMaker manages data efficiently, especially for distributed setups where data needs to be synchronized across multiple instances. - Distributed training options: Data parallelism (splitting data across instances) is common for many models, while model parallelism (splitting the model across instances) is useful for very large models that exceed instance memory.
Content from Training Models in SageMaker: PyTorch Example
Last updated on 2024-11-07 | Edit this page
Overview
Questions
- When should you consider using a GPU instance for training neural networks in SageMaker, and what are the benefits and limitations?
- How does SageMaker handle data parallelism and model parallelism, and which is suitable for typical neural network training?
Objectives
- Preprocess the Titanic dataset for efficient training using PyTorch.
- Save and upload training and validation data in
.npzformat to S3. - Understand the trade-offs between CPU and GPU training for smaller datasets.
- Deploy a PyTorch model to SageMaker and evaluate instance types for training performance.
- Differentiate between data parallelism and model parallelism, and determine when each is appropriate in SageMaker.
Initial setup
To keep things organized, you may wish to open a fresh jupyter notebook (pytorch environment). Name your notebook something along the lines of, “Training-part2.ipynb”. Once your notebook is open, we can setup our SageMaker controller as usual:
PYTHON
import boto3
import pandas as pd
import sagemaker
from sagemaker import get_execution_role
# Initialize the SageMaker role (will reflect notebook instance's policy)
role = sagemaker.get_execution_role()
print(f'role = {role}')
# Create a SageMaker session to manage interactions with Amazon SageMaker, such as training jobs, model deployments, and data input/output.
session = sagemaker.Session()
# Initialize an S3 client to interact with Amazon S3, allowing operations like uploading, downloading, and managing objects and buckets.
s3 = boto3.client('s3')
# Define the S3 bucket that we will load from
bucket_name = 'myawesometeam-titanic' # replace with your S3 bucket name
# Define train/test filenames
train_filename = 'titanic_train.csv'
test_filename = 'titanic_test.csv'We should also record our local instance information to report this information during testing.
Training a neural network with SageMaker
Let’s see how to do a similar experiment, but this time using PyTorch neural networks. We will again demonstrate how to test our custom model train script (train_nn.py) before deploying to SageMaker, and discuss some strategies (e.g., using a GPU) for improving train time when needed.
Preparing the data (compressed npz files)
When deploying a PyTorch model on SageMaker, it’s helpful to prepare the input data in a format that’s directly accessible and compatible with PyTorch’s data handling methods. The next code cell will prep our npz files from the existing csv versions.
Why are we using this file format?
Optimized data loading:
The.npzformat stores arrays in a compressed, binary format, making it efficient for both storage and loading. PyTorch can easily handle.npzfiles, especially in batch processing, without requiring complex data transformations during training.Batch compatibility:
When training neural networks in PyTorch, it’s common to load data in batches. By storing data in an.npzfile, we can quickly load the entire dataset or specific parts (e.g.,X_train,y_train) into memory and feed it to the PyTorchDataLoader, enabling efficient batched data loading.Reduced I/O overhead in SageMaker:
Storing data in.npzfiles minimizes the I/O operations during training, reducing time spent on data handling. This is especially beneficial in cloud environments like SageMaker, where efficient data handling directly impacts training costs and performance.Consistency and compatibility:
Using.npzfiles allows us to ensure consistency between training and validation datasets. Each file (train_data.npzandval_data.npz) stores the arrays in a standardized way that can be easily accessed by keys (X_train,y_train,X_val,y_val). This structure is compatible with PyTorch’sDatasetclass, making it straightforward to design custom datasets for training.Support for multiple data types:
.npzfiles support storage of multiple arrays within a single file. This is helpful for organizing features and labels without additional code. Here, thetrain_data.npzfile contains bothX_trainandy_train, keeping everything related to training data in one place. Similarly,val_data.npzorganizes validation features and labels, simplifying file management.
In summary, saving the data in .npz files ensures a
smooth workflow from data loading to model training in PyTorch,
leveraging SageMaker’s infrastructure for a more efficient, structured
training process.
PYTHON
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler, LabelEncoder
import numpy as np
# Load and preprocess the Titanic dataset
df = pd.read_csv(train_filename)
# Encode categorical variables and normalize numerical ones
df['Sex'] = LabelEncoder().fit_transform(df['Sex'])
df['Embarked'] = df['Embarked'].fillna('S') # Fill missing values in 'Embarked'
df['Embarked'] = LabelEncoder().fit_transform(df['Embarked'])
# Fill missing values for 'Age' and 'Fare' with median
df['Age'] = df['Age'].fillna(df['Age'].median())
df['Fare'] = df['Fare'].fillna(df['Fare'].median())
# Select features and target
X = df[['Pclass', 'Sex', 'Age', 'SibSp', 'Parch', 'Fare', 'Embarked']].values
y = df['Survived'].values
# Normalize features (helps avoid exploding/vanishing gradients)
scaler = StandardScaler()
X = scaler.fit_transform(X)
# Split the data
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.2, random_state=42)
# Save the preprocessed data to our local jupyter environment
np.savez('train_data.npz', X_train=X_train, y_train=y_train)
np.savez('val_data.npz', X_val=X_val, y_val=y_val)Next, we will upload our compressed files to our S3 bucket. Storage is farily cheap on AWS (around $0.023 per GB per month), but be mindful of uploading too much data. It may be convenient to store a preprocessed version of the data, just don’t store too many versions that aren’t being actively used.
PYTHON
import boto3
train_file = "train_data.npz" # Local file path in your notebook environment
val_file = "val_data.npz" # Local file path in your notebook environment
# Initialize the S3 client
s3 = boto3.client('s3')
# Upload the training and validation files to S3
s3.upload_file(train_file, bucket_name, f"{train_file}")
s3.upload_file(val_file, bucket_name, f"{val_file}")
print("Files successfully uploaded to S3.")Testing on notebook instance
You should always test code thoroughly before scaling up and using more resources. Here, we will test our script using a small number of epochs — just to verify our setup is correct.
PYTHON
import torch
import time as t # Measure training time locally
epochs = 1000
learning_rate = 0.001
start_time = t.time()
%run AWS_helpers/train_nn.py --train train_data.npz --val val_data.npz --epochs {epochs} --learning_rate {learning_rate}
print(f"Local training time: {t.time() - start_time:.2f} seconds, instance_type = {local_instance}")Deploying PyTorch neural network via SageMaker
Now that we have tested things locally, we can try to train with a
larger number of epochs and a better instance selected. We can do this
easily by invoking the PyTorch estimator. Our notebook is currently
configured to use ml.m5.large. We can upgrade this to
ml.m5.xlarge with the below code (using our notebook as a
controller).
Should we use a GPU?: Since this dataset is farily
small, we don’t necessarily need a GPU for training. Considering costs,
the m5.xlarge is $0.17/hour, while the cheapest GPU
instance is $0.75/hour. However, for larger datasets (>
1 GB) and models, we may want to consider a GPU if training time becomes
cumbersome (see Instances
for ML. If that doesn’t work, we can try distributed computing
(setting instance > 1). More on this in the next section.
PYTHON
from sagemaker.pytorch import PyTorch
from sagemaker.inputs import TrainingInput
instance_count = 1
instance_type="ml.m5.large"
output_path = f's3://{bucket_name}/output_nn/' # this folder will auto-generate if it doesn't exist already
# Define the PyTorch estimator and pass hyperparameters as arguments
pytorch_estimator = PyTorch(
entry_point="AWS_helpers/train_nn.py",
role=role,
instance_type=instance_type, # with this small dataset, we don't recessarily need a GPU for fast training.
instance_count=instance_count, # Distributed training with two instances
framework_version="1.9",
py_version="py38",
output_path=output_path,
sagemaker_session=session,
hyperparameters={
"train": "/opt/ml/input/data/train/train_data.npz", # SageMaker will mount this path
"val": "/opt/ml/input/data/val/val_data.npz", # SageMaker will mount this path
"epochs": epochs,
"learning_rate": learning_rate
}
)
# Define input paths
train_input = TrainingInput(f"s3://{bucket_name}/train_data.npz", content_type="application/x-npz")
val_input = TrainingInput(f"s3://{bucket_name}/val_data.npz", content_type="application/x-npz")
# Start the training job and time it
start = t.time()
pytorch_estimator.fit({"train": train_input, "val": val_input})
end = t.time()
print(f"Runtime for training on SageMaker: {end - start:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")Deploying PyTorch neural network via SageMaker with a GPU instance
In this section, we’ll implement the same procedure as above, but using a GPU-enabled instance for potentially faster training. While GPU instances are more expensive, they can be cost-effective for larger datasets or more complex models that require significant computational power.
Selecting a GPU Instance
For a small dataset like ours, we don’t strictly need a GPU, but for
larger datasets or more complex models, a GPU can reduce training time.
Here, we’ll select an ml.g4dn.xlarge instance, which
provides a single GPU and costs approximately $0.75/hour
(check Instances
for ML for detailed pricing).
Code modifications for GPU use
Using a GPU requires minor changes in your training script
(train_nn.py). Specifically, you’ll need to: 1. Check for
GPU availability in PyTorch. 2. Move the model and tensors to the GPU
device if available.
Enabling PyTorch to use GPU in train_nn.py
The following code snippet to enables GPU support in
train_nn.py:
PYTHON
import torch
# Set device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")PYTHON
from sagemaker.pytorch import PyTorch
from sagemaker.inputs import TrainingInput
import time as t
instance_count = 1
instance_type="ml.g4dn.xlarge"
output_path = f's3://{bucket_name}/output_nn/'
# Define the PyTorch estimator and pass hyperparameters as arguments
pytorch_estimator_gpu = PyTorch(
entry_point="AWS_helpers/train_nn.py",
role=role,
instance_type=instance_type,
instance_count=instance_count,
framework_version="1.9",
py_version="py38",
output_path=output_path,
sagemaker_session=session,
hyperparameters={
"train": "/opt/ml/input/data/train/train_data.npz",
"val": "/opt/ml/input/data/val/val_data.npz",
"epochs": epochs,
"learning_rate": learning_rate
}
)
# Define input paths
train_input = TrainingInput(f"s3://{bucket_name}/train_data.npz", content_type="application/x-npz")
val_input = TrainingInput(f"s3://{bucket_name}/val_data.npz", content_type="application/x-npz")
# Start the training job and time it
start = t.time()
pytorch_estimator_gpu.fit({"train": train_input, "val": val_input})
end = t.time()
print(f"Runtime for training on SageMaker: {end - start:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")SH
2024-11-03 21:33:56 Uploading - Uploading generated training model
2024-11-03 21:33:56 Completed - Training job completed
Training seconds: 350
Billable seconds: 350
Runtime for training on SageMaker: 409.68 seconds, instance_type: ml.g4dn.xlarge, instance_count: 1GPUs can be slow for small datasets/models
This performance discrepancy might be due to the following factors:
Small Ddtaset/model size: When datasets and models are small, the overhead of transferring data between the CPU and GPU, as well as managing the GPU, can actually slow things down. For very small models and datasets, CPUs are often faster since there’s minimal data to process.
GPU initialization overhead: Every time a training job starts on a GPU, there’s a small overhead for initializing CUDA libraries. For short jobs, this setup time can make the GPU appear slower overall.
Batch size: GPUs perform best with larger batch sizes since they can process many data points in parallel. If the batch size is too small, the GPU is underutilized, leading to suboptimal performance. You may want to try increasing the batch size to see if this reduces training time.
Instance type: Some GPU instances, like the
ml.g4dnseries, have less computational power than the largerp3series. They’re better suited for inference or lightweight tasks rather than intense training, so a more powerful instance (e.g.,ml.p3.2xlarge) could help for larger tasks.
If training time continues to be critical, sticking with a CPU instance may be the best approach for smaller datasets. For larger, more complex models and datasets, the GPU’s advantages should become more apparent.
Distributed Training for Neural Networks in SageMaker
In the event that you do need distributed computing to achieve reasonable train times (remember to try an upgraded instance first!), simply adjust the instance count to a number between 2 and 5. Beyond 5 instances, you’ll see diminishing returns and may be needlessly spending extra money/compute-energy.
PYTHON
from sagemaker.pytorch import PyTorch
from sagemaker.inputs import TrainingInput
import time as t
instance_count = 2 # increasing to 2 to see if it has any benefit (likely won't see any with this small dataset)
instance_type="ml.m5.xlarge"
output_path = f's3://{bucket_name}/output_nn/'
# Define the PyTorch estimator and pass hyperparameters as arguments
pytorch_estimator = PyTorch(
entry_point="AWS_helpers/train_nn.py",
role=role,
instance_type=instance_type, # with this small dataset, we don't recessarily need a GPU for fast training.
instance_count=instance_count, # Distributed training with two instances
framework_version="1.9",
py_version="py38",
output_path=output_path,
sagemaker_session=session,
hyperparameters={
"train": "/opt/ml/input/data/train/train_data.npz", # SageMaker will mount this path
"val": "/opt/ml/input/data/val/val_data.npz", # SageMaker will mount this path
"epochs": epochs,
"learning_rate": learning_rate
}
)
# Define input paths
train_input = TrainingInput(f"s3://{bucket_name}/train_data.npz", content_type="application/x-npz")
val_input = TrainingInput(f"s3://{bucket_name}/val_data.npz", content_type="application/x-npz")
# Start the training job and time it
start = t.time()
pytorch_estimator.fit({"train": train_input, "val": val_input})
end = t.time()
print(f"Runtime for training on SageMaker: {end - start:.2f} seconds, instance_type: {instance_type}, instance_count: {instance_count}")SH
2024-11-03 21:36:35 Uploading - Uploading generated training model
2024-11-03 21:36:47 Completed - Training job completed
Training seconds: 228
Billable seconds: 228
Runtime for training on SageMaker: 198.36 seconds, instance_type: ml.m5.xlarge, instance_count: 2Distributed training for neural nets: how epochs are managed
Amazon SageMaker provides two main strategies for distributed training: data parallelism and model parallelism. Understanding which strategy will be used depends on the model size and the configuration of your SageMaker training job, as well as the default settings of the specific SageMaker Estimator you are using.
1. Data parallelism (most common for mini-batch SGD)
-
How it works: In data parallelism, each instance in
the cluster (e.g., multiple
ml.m5.xlargeinstances) maintains a complete copy of the model. The training dataset is split across instances, and each instance processes a different subset of data simultaneously. This enables multiple instances to complete forward and backward passes on different data batches independently. - Epoch distribution: Even though each instance processes all the specified epochs, they only work on a portion of the dataset for each epoch. After each batch, instances synchronize their gradient updates across all instances using a method such as all-reduce. This ensures that while each instance is working with a unique data batch, the model weights remain consistent across instances.
- Key insight: Because all instances process the specified number of epochs and synchronize weight updates between batches, each instance’s training contributes to a cohesive, shared model. The effective epoch count across instances appears to be shared because data parallelism allows each instance to handle a fraction of the data per epoch, not the epochs themselves. Data parallelism is well-suited for models that can fit into a single instance’s memory and benefit from increased data throughput.
2. Model parallelism (best for large models)
- How it works: Model parallelism divides the model itself across multiple instances, not the data. This approach is best suited for very large models that cannot fit into a single GPU or instance’s memory (e.g., large language models).
- Epoch distribution: The model is partitioned so that each instance is responsible for specific layers or components. Data flows sequentially through these partitions, where each instance processes a part of each batch and passes it to the next instance.
- Key insight: This approach is more complex due to the dependency between model components, so synchronization occurs across the model layers rather than across data batches. Model parallelism generally suits scenarios with exceptionally large model architectures that exceed memory limits of typical instances.
Determining which distributed training strategy is used
SageMaker will select the distributed strategy based on: -
Framework and Estimator configuration: Most deep
learning frameworks in SageMaker default to data parallelism, especially
when using PyTorch or TensorFlow with standard configurations. -
Model and data size: If you specify a model that
exceeds a single instance’s memory capacity, SageMaker may switch to
model parallelism if configured for it. - Instance
count: When you specify instance_count > 1 in
your Estimator with a deep learning model, SageMaker will use data
parallelism by default unless explicitly configured for model
parallelism.
You observed that each instance ran all epochs with
instance_count=2 and 10,000 epochs, which aligns with data
parallelism. Here, each instance processed the full set of epochs
independently, but each batch of data was different, and the gradient
updates were synchronized across instances.
Key Points
-
Efficient data handling: The
.npzformat is optimized for efficient loading, reducing I/O overhead and enabling batch compatibility for PyTorch’sDataLoader. - GPU training: While beneficial for larger models or datasets, GPUs may introduce overhead for smaller tasks; selecting the right instance type is critical for cost-efficiency.
- Data parallelism vs. model parallelism: Data parallelism splits data across instances and synchronizes model weights, suitable for typical neural network tasks. Model parallelism, which divides model layers, is ideal for very large models that exceed memory capacity.
- SageMaker configuration: By adjusting instance counts and types, SageMaker supports scalable training setups. Starting with CPU training and scaling as needed with GPUs or distributed setups allows for performance optimization.
- Testing locally first: Before deploying large-scale training in SageMaker, test locally with a smaller setup to ensure code correctness and efficient resource usage.
Content from Hyperparameter Tuning in SageMaker: Neural Network Example
Last updated on 2024-11-07 | Edit this page
Overview
Questions
- How can we efficiently manage hyperparameter tuning in SageMaker?
- How can we parallelize tuning jobs to optimize time without increasing costs?
Objectives
- Set up and run a hyperparameter tuning job in SageMaker.
- Define
ContinuousParameterandCategoricalParameterfor targeted tuning. - Log and capture objective metrics for evaluating tuning success.
- Optimize tuning setup to balance cost and efficiency, including parallelization.
To conduct efficient hyperparameter tuning with neural networks (or any model) in SageMaker, we’ll leverage SageMaker’s hyperparameter tuning jobs while carefully managing parameter ranges and model count. Here’s an overview of the process, with a focus on both efficiency and cost-effectiveness.
Key steps for hyperparameter tuning
The overall process involves these five below steps.
- Setup estimator
- Define parameter ranges
- Set up HyperParamterTuner object
- Prepare training script to log metrics
- Set data paths and launch tuner.fit()
- Monitor tuning job from SageMaker console
- Extract best model for final evaluation
Code example for SageMaker hyperparameter tuning with neural networks
We’ll walk through each step in detail by tuning a neural network.
Specifcially, we will test out different values for our
epochs and learning_rate parameters. We are
sticking to just two hyperparameters for demonstration purposes, but you
may wish to explore additional parameters in your own work.
This setup provides: - Explicit control over
epochs using CategoricalParameter, allowing
targeted testing of specific values. - Efficient
sampling for learning_rate using
ContinuousParameter, covering a defined range for a
balanced approach. - Cost control by setting moderate
max_jobs and max_parallel_jobs.
By managing these settings and configuring metrics properly, you can achieve a balanced and efficient approach to hyperparameter tuning for neural networks.
0. Directory setup
Just to make we are all on the same directory starting point, let’s cd to our instance’s root directory
1. Setup estimator
To kick off our hyperparameter tuning for a neural network model, we’ll start by defining the SageMaker Estimator. The estimator setup here is very similar to our previous episode, where we used it to configure and train a model directly. However, this time, rather than directly running a training job with the estimator, we’ll be using it as the foundation for hyperparameter tuning.
In SageMaker, the estimator serves as a blueprint for each tuning job, specifying the training script, instance type, and key parameters like data paths and metrics. Once defined, this estimator will be passed to a Hyperparameter Tuner that manages the creation of multiple training jobs with various hyperparameter combinations, helping us find an optimal configuration.
Here’s the setup for our PyTorch estimator, which includes specifying
the entry script for training (train_nn.py) and defining
hyperparameters that will remain fixed across tuning jobs. The
hyperparameters we’re setting up to tune include epochs and
learning_rate, with a few specific values or ranges
defined:
PYTHON
import sagemaker
from sagemaker.tuner import HyperparameterTuner, IntegerParameter, ContinuousParameter, CategoricalParameter
from sagemaker.pytorch import PyTorch
from sagemaker.inputs import TrainingInput
from sagemaker import get_execution_role
# Initialize SageMaker session and role
session = sagemaker.Session()
role = get_execution_role()
bucket_name = 'myawesometeam-titanic' # replace with your S3 bucket name
# Define the PyTorch estimator with entry script and environment details
pytorch_estimator = PyTorch(
entry_point="AWS_helpers/train_nn.py", # Your script for training
role=role,
instance_count=1,
instance_type="ml.m5.large",
framework_version="1.9",
py_version="py38",
metric_definitions=[{"Name": "validation:accuracy", "Regex": "validation:accuracy = ([0-9\\.]+)"}],
hyperparameters={
"train": "/opt/ml/input/data/train/train_data.npz", # SageMaker will mount this path
"val": "/opt/ml/input/data/val/val_data.npz", # SageMaker will mount this path
"epochs": 100, # Placeholder initial value. Will be overridden by tuning by tuning values tested
"learning_rate": 0.001 # Placeholder initial value. Will be overridden by tuning values tested
},
sagemaker_session=session,
)
2. Define hyperparameter ranges
In SageMaker, you must explicitly define ranges for any
hyperparameters you want to tune. SageMaker supports both
ContinuousParameter and CategoricalParameter
types:
-
ContinuousParameterallows SageMaker to dynamically sample numeric values within a specified range, making it ideal for broad, exploratory tuning. The total number of values tested can be controlled through the upcomingmax_jobsparameter, which defines how many different combinations SageMaker will evaluate. -
CategoricalParameterspecifies exact values for SageMaker to test, which is useful when you want the model to only try a specific set of options.
By default, SageMaker uses Bayesian optimization,
adjusting future selections based on previous results to efficiently
find optimal values. You can also set the strategy to
“Random” for uniform sampling across the range, which
is effective in larger or more exploratory search spaces. Random
sampling may end up costing much more in time and resources, however.
Generally, we recommend sticking with the default setting.
Hyperparameter considerations in neural nets
When tuning hyperparameters in neural networks, it’s essential to prioritize parameters that directly impact model performance while remaining mindful of diminishing returns and potential overfitting. Below are some core hyperparameters to consider and general strategies for tuning:
Learning Rate: Often the most impactful parameter, learning rate controls the speed of convergence. A smaller learning rate can lead to more stable, though slower, convergence, while a larger rate may speed things up but risks overshooting optimal values. Testing ranges like
0.0001to0.1with aContinuousParameteris common practice, andBayesiansearch can help home in on ideal values.Batch Size: Larger batch sizes often yield faster training times and may improve stability, but this can risk bypassing useful local minima, especially with small datasets. Smaller batch sizes can capture more nuanced updates but are computationally expensive. Ranges from
16to256are worth exploring for most use cases, although, for large datasets or high-capacity models, even larger values may be beneficial.Number of Epochs: While larger epochs allow the model to learn from data longer, increasing epochs doesn’t always equate to better performance and can lead to overfitting. Exploring
CategoricalParameter([50, 100, 500, 1000])can help balance performance without excessive training costs.Layer Width and Depth: Increasing the width or depth (i.e., number of neurons and layers) can improve model capacity but also risks overfitting if the dataset is small or lacks variability. Testing a range of layer sizes or depths (e.g.,
32, 64, 128neurons per layer) can reveal whether additional complexity yields benefits. Notably, understanding double descent is essential here, as larger networks may initially seem to overfit before unexpectedly improving in the second descent—a phenomenon worth investigating in high-capacity networks.Regularization Parameters: Regularization techniques, such as dropout rates or weight decay, can help control overfitting by limiting model capacity. For example, dropout rates from
0.1to0.5or weight decay values of0.0001to0.01often balance underfitting and overfitting effectively. Higher regularization might inhibit learning, especially if the model is relatively small.Early Stopping: While not a traditional hyperparameter, setting up early stopping based on the validation performance can prevent overfitting without the need to exhaustively test for epoch limits. By allowing the model to halt once performance plateaus or worsens, early stopping can improve efficiency in hyperparameter tuning.
Special Phenomena - Grokking and Double Descent: For certain complex datasets or when tuning particularly large models, keep an eye on phenomena like grokking (sudden shifts from poor to excellent generalization) and double descent (an unexpected second drop in error after initial overfitting). These behaviors are more likely to appear in models with high capacity and prolonged training periods, potentially requiring longer epochs or lower learning rates to observe.
In summary, hyperparameter tuning is a balancing act between expanding model capacity and mitigating overfitting. Prioritize parameters that have shown past efficacy in similar problems, and limit the search to a manageable range—often 20–30 model configurations are sufficient to observe gains. This approach keeps resource consumption practical while achieving meaningful improvements in model performance.
3. Set up HyperParamterTuner object
In step 3, we set up the HyperparameterTuner, which
controls the tuning process by specifying the…
-
estimator: Here, we connect the previously definedpytorch_estimatortotuner, ensuring that the tuning job will run with our PyTorch model configuration. -
objectives: - The
metric_definitionsandobjective_metric_nameindicate which metric SageMaker should monitor to find the best-performing model; in this case, we’re tracking “validation:accuracy” and aiming to maximize it. We’ll show you how to setup your training script to report this information in the next step. -
hyperparameter ranges: Defined above. -
tuning strategy: SageMaker uses a Bayesian strategy by default, which iteratively improves parameter selection based on past performance to find an optimal model more efficiently. Although it’s possible to adjust to a “Random” strategy, Bayesian optimization generally provides better results, so it’s recommended to keep this setting. -
max_jobsandmax_parallel_jobs: Finally, we setmax_jobsto control the total number of configurations SageMaker will explore andmax_parallel_jobsto limit the number of jobs that run simultaneously, balancing resource usage and tuning speed. Since SageMaker tests different hyperparameter values dynamically, it’s important to limit total parallel instances to <= 4.
Resource-Conscious Approach: To control costs and energy-needs, choose efficient instance types and limit the search to impactful parameters at a sensible range, keeping resource consumption in check. Hyperparameter tuning often does yield better performing models, but these gains can be marginal after exhausting a reasonable search window of 20-30 model configurations. As a researcher, it’s also imortant to do some digging on past work to see which parameters may be worthwhile to explore. Make sure you understand what each parameter is doing before you adjust them.
Always start with max_jobs = 1 and
max_parallel_jobs=1. Before running the full
search, let’s test our setup by setting max_jobs = 1. This will test
just one possible hyperparameter configuration. This critical step helps
ensure our code is functional before attempting to scale up.
PYTHON
# Tuner configuration
tuner = HyperparameterTuner(
estimator=pytorch_estimator,
metric_definitions=[{"Name": "validation:accuracy", "Regex": "validation:accuracy = ([0-9\\.]+)"}],
objective_metric_name="validation:accuracy", # Ensure this matches the metric name exactly
objective_type="Maximize", # Specify if maximizing or minimizing the metric
hyperparameter_ranges=hyperparameter_ranges,
strategy="Bayesian", # Default setting (recommend sticking with this!); can adjust to "Random" for uniform sampling across the range
max_jobs=1, # Always start with 1 instance for debugging purposes. Adjust based on exploration needs (keep below 30 to be kind to environment).
max_parallel_jobs=1 # Always start with 1 instance for debugging purposes. Adjust based on available resources and budget. Recommended to keep this value < 4 since SageMaker tests values dynamically.
)4. Prepare training script to log metrics
To prepare train_nn.py for hyperparameter tuning, we
added code to log validation metrics in a format that SageMaker
recognizes for tracking. In the training loop, we added a print
statement for Val Accuracy in a specific format that
SageMaker can capture.
Note: It’s best to use an if statement to only print out metrics periodically (e.g., every 100 epochs), so that you print time does not each up too much of your training time. It may be a little counter-intuitive that printing can slow things down so dramatically, but it truly does become a significant factor if you’re doing it every epoch. On the flipside of this, you don’t want to print metrics so infrequently that you lose resolution in the monitored validation accuracy. Choose a number between 100-1000 epochs or divide your total epoch count by ~25 to yield a reasonable range.
PYTHON
if (epoch + 1) % 100 == 0 or epoch == epochs - 1:
print(f"validation:accuracy = {val_accuracy:.4f}", flush=True) # Log for SageMaker metric tracking. Needed for hyperparameter tuning later.Paired with this, our metric_definitions above uses a
regular expression "validation:accuracy = ([0-9\\.]+)" to
extract the val_accuracy value from each log line. This regex
specifically looks for validation:accuracy =, followed by a
floating-point number, which corresponds to the format of our log
statement in train_nn.py.
5. Set data paths and launch tuner.fit()
In step 4, we define the input data paths for the training job and
launch the hyperparameter tuning process. Using
TrainingInput, we specify the S3 paths to our
train_data.npz and val_data.npz files. This
setup ensures that SageMaker correctly accesses our training and
validation data during each job in the tuning process. We then call
tuner.fit and pass a dictionary mapping each data channel
(“train” and “val”) to its respective path. This command initiates the
tuning job, triggering SageMaker to begin sampling hyperparameter
combinations, training the model, and evaluating performance based on
our defined objective metric. Once the job is launched, SageMaker
handles the tuning process automatically, running the specified number
of configurations and keeping track of the best model parameters based
on validation accuracy.
PYTHON
# Define the input paths
train_input = TrainingInput(f"s3://{bucket_name}/train_data.npz", content_type="application/x-npz")
val_input = TrainingInput(f"s3://{bucket_name}/val_data.npz", content_type="application/x-npz")
# Launch the hyperparameter tuning job
tuner.fit({"train": train_input, "val": val_input})
print("Hyperparameter tuning job launched.")SH
No finished training job found associated with this estimator. Please make sure this estimator is only used for building workflow config
No finished training job found associated with this estimator. Please make sure this estimator is only used for building workflow config
.......................................!
Hyperparameter tuning job launched.Scaling up our approach
If all goes well, we can scale up the experiment with the below code.
In this configuration, we’re scaling up the search by allowing SageMaker
to test more hyperparameter configurations (max_jobs=20)
while setting max_parallel_jobs=2 to manage parallelization
efficiently. With two jobs running at once, SageMaker will be able to
explore potential improvements more quickly than in a fully sequential
setup, while still dynamically selecting promising values as it learns
from completed jobs. This balance leverages SageMaker’s Bayesian
optimization, which uses completed trials to inform subsequent ones,
helping to avoid redundant testing of less promising parameter
combinations. Setting max_parallel_jobs higher than
2-4 could increase costs and reduce tuning efficiency, as SageMaker’s
ability to learn from completed jobs decreases when too many jobs run
simultaneously.
With this approach, SageMaker is better able to refine configurations
without overloading resources or risking inefficient exploration, making
max_parallel_jobs=2 a solid default for most use cases.
PYTHON
import time as t # always a good idea to keep a runtime of your experiments
# Configuration variables
instance_type = "ml.m5.large"
max_jobs = 2
max_parallel_jobs = 2
# Define the Tuner configuration
tuner = HyperparameterTuner(
estimator=pytorch_estimator,
metric_definitions=[{"Name": "validation:accuracy", "Regex": "validation:accuracy = ([0-9\\.]+)"}],
objective_metric_name="validation:accuracy", # Ensure this matches the metric name exactly
objective_type="Maximize", # Specify if maximizing or minimizing the metric
hyperparameter_ranges=hyperparameter_ranges,
max_jobs=max_jobs,
max_parallel_jobs=max_parallel_jobs
)
# Define the input paths
train_input = TrainingInput(f"s3://{bucket_name}/train_data.npz", content_type="application/x-npz")
val_input = TrainingInput(f"s3://{bucket_name}/val_data.npz", content_type="application/x-npz")
# Track start time
start_time = t.time()
# Launch the hyperparameter tuning job
tuner.fit({"train": train_input, "val": val_input})
# Calculate runtime
runtime = t.time() - start_time
# Print confirmation with runtime and configuration details
print(f"Tuning runtime: {runtime:.2f} seconds, Instance Type: {instance_type}, Max Jobs: {max_jobs}, Max Parallel Jobs: {max_parallel_jobs}")SH
No finished training job found associated with this estimator. Please make sure this estimator is only used for building workflow config
No finished training job found associated with this estimator. Please make sure this estimator is only used for building workflow config
.......................................!
Tuning runtime: 205.53 seconds, Instance Type: ml.m5.large, Max Jobs: 2, Max Parallel Jobs: 2Monitoring tuning
After running the above cell, we can check on the progress by visiting the SageMaker Console and finding the “Training” tab located on the left panel. Click “Hyperparmater tuning jobs” to view running jobs.
- Initial Jobs: SageMaker starts by running only max_parallel_jobs (2 in this case) as the initial batch. As each job completes, new jobs from the remaining pool are triggered until max_jobs (20) is reached.
- Job Completion: Once the first few jobs complete, SageMaker will continue to launch the remaining jobs up to the maximum of 20, but no more than two at a time.
Can/should we run more instances in parallel?
Setting max_parallel_jobs to 20 (equal to max_jobs) will indeed launch all 20 jobs in parallel. This approach won’t affect the total cost (since cost is based on the number of total jobs, not how many run concurrently), but it can impact the final results and resource usage pattern due to SageMaker’s ability to dynamically select hyperparameter values to test to maximize efficiency and improve model performance. This adaptability is especially useful for neural networks, which often have a large hyperparameter space with complex interactions. Here’s how SageMaker’s approach impacts typical neural network training:
1. Adaptive Search Strategies
- SageMaker offers Bayesian optimization for hyperparameter tuning. Instead of purely random sampling, it learns from previous jobs to choose the next set of hyperparameters more likely to improve the objective metric.
- For neural networks, this strategy can help converge on better-performing configurations faster by favoring promising areas of the hyperparameter space and discarding poor ones.
2. Effect of max_parallel_jobs on adaptive tuning
- When using Bayesian optimization, a lower
max_parallel_jobs(e.g., 2–4) can allow SageMaker to iteratively adjust and improve its choices. Each batch of jobs informs the subsequent batch, which may yield better results over time. - Conversely, if all jobs are run in parallel (e.g.,
max_parallel_jobs=20), SageMaker can’t learn and adapt within a single batch, making this setup more like a traditional grid or random search. This approach is still valid, especially for small search spaces, but it doesn’t leverage the full potential of adaptive tuning.
3. Practical impact on neural network training
-
For simpler models or smaller parameter ranges,
running jobs in parallel with a higher
max_parallel_jobsworks well and quickly completes the search. -
For more complex neural networks or large
hyperparameter spaces, an adaptive strategy with a smaller
max_parallel_jobsmay yield a better model with fewer total jobs by fine-tuning hyperparameters over multiple iterations.
Summary
-
For fast, straightforward tuning: Set
max_parallel_jobscloser tomax_jobsfor simultaneous testing. -
For adaptive, refined tuning: Use a smaller
max_parallel_jobs(like 2–4) to let SageMaker leverage adaptive tuning for optimal configurations.
This balance between exploration and exploitation is particularly impactful in neural network tuning, where training costs can be high and parameters interact in complex ways.
Extracting and evaluating the best model after tuning
Tuning should only take about 5 minutes to complete — not bad for 20 models! After SageMaker completes the hyperparameter tuning job, the results, including the trained models for each configuration, are stored in an S3 bucket. Here’s a breakdown of the steps to locate and evaluate the best model on test data.
-
Understanding the folder structure:
- SageMaker saves each tuning job’s results in the specified S3 bucket under a unique prefix.
- For the best model, SageMaker stores the model artifact in the
format
s3://{bucket}/{job_name}/output/model.tar.gz. Each model is compressed as a.tar.gzfile containing the saved model parameters.
-
Retrieve and load the best model:
- Using the
tuner.best_training_job()method, you can get the name of the best-performing job. - From there, retrieve the S3 URI of the best model artifact, download it locally, and extract the files for use.
- Using the
-
Prepare test data for final assessment of model
generalizability
- If not done already.
-
Evaluate the model on test data:
- Once extracted, load the saved model weights and evaluate the model on your test dataset to get the final performance metrics.
Here’s the code to implement these steps:
View best model details and storage info
We can easily view the best hyperparameters from the tuning procedure…
PYTHON
# 1. Get the best training job from the completed tuning job
best_job_name = tuner.best_training_job()
print("Best training job name:", best_job_name)
# 2. Use describe_training_job to retrieve full details, including hyperparameters...
best_job_desc = session.sagemaker_client.describe_training_job(TrainingJobName=best_job_name)
best_hyperparameters = best_job_desc["HyperParameters"]
print("Best hyperparameters:", best_hyperparameters)
# ... and model URI (location on S3)
best_model_s3_uri = best_job_desc['ModelArtifacts']['S3ModelArtifacts']
print(f"Best model artifact S3 URI: {best_model_s3_uri}")SH
Best training job name: pytorch-training-241107-0025-001-72851d7f
Best hyperparameters: {'_tuning_objective_metric': 'validation:accuracy', 'epochs': '"100"', 'learning_rate': '0.005250489250786233', 'sagemaker_container_log_level': '20', 'sagemaker_estimator_class_name': '"PyTorch"', 'sagemaker_estimator_module': '"sagemaker.pytorch.estimator"', 'sagemaker_job_name': '"pytorch-training-2024-11-07-00-25-35-999"', 'sagemaker_program': '"train_nn.py"', 'sagemaker_region': '"us-east-1"', 'sagemaker_submit_directory': '"s3://sagemaker-us-east-1-183295408236/pytorch-training-2024-11-07-00-25-35-999/source/sourcedir.tar.gz"', 'train': '"/opt/ml/input/data/train/train_data.npz"', 'val': '"/opt/ml/input/data/val/val_data.npz"'}
Best model artifact S3 URI: s3://sagemaker-us-east-1-183295408236/pytorch-training-241107-0025-001-72851d7f/output/model.tar.gzRetrieve and load best model
PYTHON
import boto3
import tarfile
# Initialize S3 client
s3 = boto3.client('s3')
# Download and extract the model artifact
local_model_path = "best_model.tar.gz"
bucket_name, model_key = best_model_s3_uri.split('/')[2], '/'.join(best_model_s3_uri.split('/')[3:])
s3.download_file(bucket_name, model_key, local_model_path)
# Extract the model files from the tar.gz archive
with tarfile.open(local_model_path, 'r:gz') as tar:
tar.extractall()
print("Best model downloaded and extracted.")Prepare test set as test_data.npz
In our previous episode, we converted our train dataset into train/validate subsets, and saved them out as .npz files for efficient processing. We’ll need to preprocess our test data the same way to evaluate it on our model.
Note: It’s always a good idea to keep preprocessing
code as a function so you can apply the same exact procedure across
datasets with ease. We’ll import our preprocessing function from
train_nn.py.
PYTHON
import pandas as pd
import numpy as np
# Now try importing the function again
from AWS_helpers.train_nn import preprocess_data
# Example usage for test data (using the same scaler from training)
test_df = pd.read_csv("titanic_test.csv")
X_test, y_test, _ = preprocess_data(test_df)
# Save processed data for testing
np.savez('test_data.npz', X_test=X_test, y_test=y_test)Evaluate the model on test data
PYTHON
from AWS_helpers.train_nn import TitanicNet
from AWS_helpers.train_nn import calculate_accuracy
import torch
# Load the model (assuming it's saved as 'nn_model.pth' after extraction)
model = TitanicNet() # Ensure TitanicNet is defined as per your training script
model.load_state_dict(torch.load("nn_model.pth"))
model.eval()
# Load test data (assuming the test set is saved as "test_data.npz" in npz format)
test_data = np.load("test_data.npz") # Replace "test_data.npz" with actual test data path if different
X_test = torch.tensor(test_data['X_test'], dtype=torch.float32)
y_test = torch.tensor(test_data['y_test'], dtype=torch.float32)
# Evaluate the model on the test set
with torch.no_grad():
predictions = model(X_test)
accuracy = calculate_accuracy(predictions, y_test) # Assuming calculate_accuracy is defined as in your training script
print(f"Test Accuracy: {accuracy:.4f}")Conclusions
In just under 5 minutes, we produced a model that is almost 100% accurate on the test set. However, this performance does come at a cost (albeit manageable if you’ve stuck with our advise thus far). This next section will help you assess the total compute time that was used by your tuning job.
In SageMaker, training time and billing time (extracted in our code below) are often expected to differ slightly for training jobs, but not for tuning jobs. Here’s a breakdown of what’s happening:
Training Time: This is the actual wall-clock time that each training job takes to run, from start to end. This metric represents the pure time spent on training without considering the compute resources.
Billing Time: This includes the training time but is adjusted for the resources used. Billing time considers:
Instance Count: The number of instances used for training affects billing time. Round-Up Policy: SageMaker rounds up the billing time to the nearest second for each job and multiplies it by the instance count used. This means that for short jobs, the difference between training and billing time can be more pronounced.
PYTHON
import boto3
import math
# Initialize SageMaker client
sagemaker_client = boto3.client("sagemaker")
# Retrieve tuning job details
tuning_job_name = tuner.latest_tuning_job.name # Replace with your tuning job name if needed
tuning_job_desc = sagemaker_client.describe_hyper_parameter_tuning_job(HyperParameterTuningJobName=tuning_job_name)
# Extract relevant settings
instance_type = tuning_job_desc['TrainingJobDefinition']['ResourceConfig']['InstanceType']
max_jobs = tuning_job_desc['HyperParameterTuningJobConfig']['ResourceLimits']['MaxNumberOfTrainingJobs']
max_parallel_jobs = tuning_job_desc['HyperParameterTuningJobConfig']['ResourceLimits']['MaxParallelTrainingJobs']
# Retrieve all training jobs for the tuning job
training_jobs = sagemaker_client.list_training_jobs_for_hyper_parameter_tuning_job(
HyperParameterTuningJobName=tuning_job_name, StatusEquals='Completed'
)["TrainingJobSummaries"]
# Calculate total training and billing time
total_training_time = 0
total_billing_time = 0
for job in training_jobs:
job_name = job["TrainingJobName"]
job_desc = sagemaker_client.describe_training_job(TrainingJobName=job_name)
# Calculate training time (in seconds)
training_time = job_desc["TrainingEndTime"] - job_desc["TrainingStartTime"]
total_training_time += training_time.total_seconds()
# Calculate billed time with rounding up
billed_time = math.ceil(training_time.total_seconds())
total_billing_time += billed_time * job_desc["ResourceConfig"]["InstanceCount"]
# Print configuration details and total compute/billing time
print(f"Instance Type: {instance_type}")
print(f"Max Jobs: {max_jobs}")
print(f"Max Parallel Jobs: {max_parallel_jobs}")
print(f"Total training time across all jobs: {total_training_time / 3600:.2f} hours")
print(f"Estimated total billing time across all jobs: {total_billing_time / 3600:.2f} hours")SH
Instance Type: ml.m5.large
Max Jobs: 2
Max Parallel Jobs: 2
Total training time across all jobs: 0.07 hours
Estimated total billing time across all jobs: 0.07 hoursFor convenience, we have added this as a function in helpers.py
PYTHON
import AWS_helpers.helpers as helpers
import importlib
importlib.reload(helpers)
helpers.calculate_tuning_job_time(tuner)SH
Instance Type: ml.m5.large
Max Jobs: 2
Max Parallel Jobs: 2
Total training time across all jobs: 0.07 hours
Estimated total billing time across all jobs: 0.07 hoursSH
[NbConvertApp] Converting notebook Hyperparameter-tuning.ipynb to markdown
[NbConvertApp] Writing 31418 bytes to Hyperparameter-tuning.mdPYTHON
Content from Resource Management and Monitoring
Last updated on 2024-11-08 | Edit this page
Overview
Questions
- How can I monitor and manage AWS resources to avoid unnecessary costs?
- What steps are necessary to clean up SageMaker and S3 resources after the workshop?
- What best practices can help with efficient resource utilization?
Objectives
- Understand how to shut down SageMaker notebook instances to minimize costs.
- Learn to clean up S3 storage and terminate unused training jobs.
- Explore basic resource management strategies and tools for AWS.
Shutting down notebook instances
Notebook instances in SageMaker are billed per hour, so it’s
essential to stop or delete them when they are no longer needed. Earlier
in the Notebooks as controllers episode, we discussed
using lower-cost instance types like ml.t3.medium
(approximately $0.05/hour) for controlling workflows. While this makes
open notebooks less costly than larger instances, it’s still a good
habit to stop or delete notebooks to avoid unnecessary spending,
especially if left idle for long periods.
- Navigate to SageMaker in the AWS Console.
- In the left-hand menu, click Notebooks.
- Locate your notebook instance and select it.
- Choose Stop to shut it down temporarily or Delete to permanently remove it. > Tip: If you plan to reuse the notebook later, stopping it is sufficient. Deleting is recommended if you are finished with the workshop.
Cleaning up S3 storage
While S3 storage is relatively inexpensive, cleaning up unused buckets and files helps keep costs minimal and your workspace organized.
- Navigate to the S3 Console.
- Locate the bucket(s) you created for this workshop.
- Open the bucket and select any objects (files) you no longer need.
- Click Delete to remove the selected objects.
- To delete an entire bucket:
- Empty the bucket by selecting Empty bucket under Bucket actions.
- Delete the bucket by clicking Delete bucket.
Reminder: Earlier in the workshop, we set up tags for S3 buckets. Use these tags to filter and identify workshop-related buckets, ensuring that only unnecessary resources are deleted.
Monitoring and stopping active jobs
SageMaker charges for training and tuning jobs while they run, so make sure to terminate unused jobs.
- In the SageMaker Console, go to Training Jobs or Tuning Jobs.
- Identify any active jobs that you no longer need.
- Select the jobs and click Stop. > Tip: Review the job logs to ensure you’ve saved the results before stopping a job.
Billing and cost monitoring
Managing your AWS expenses is vital to staying within budget. Follow these steps to monitor and control costs:
-
Set up billing alerts:
- Go to the AWS Billing Dashboard.
- Navigate to Budgets and create a budget alert to track your spending.
-
Review usage and costs:
- Use the AWS Cost Explorer in the Billing Dashboard to view detailed expenses by service, such as SageMaker and S3.
-
Use tags for cost tracking:
- Refer to the tags you set up earlier in the workshop for your notebooks and S3 buckets. These tags help you identify and monitor costs associated with specific resources.
Best practices for resource management
Efficient resource management can save significant costs and improve your workflows. Below are some best practices:
- Automate resource cleanup: Use AWS SDK or CLI scripts to automatically shut down instances and clean up S3 buckets when not in use. Learn more about automation with AWS CLI.
- Schedule resource usage: Schedule instance start and stop times using AWS Lambda. Learn how to schedule tasks with AWS Lambda.
- Test workflows locally first: Before scaling up experiments in SageMaker, test them locally to minimize cloud usage and costs. Learn about SageMaker local mode.
- Use cost tracking tools: Explore AWS’s cost allocation tags and budget tracking features. Learn more about cost management in AWS.
By following these practices and leveraging the additional resources provided, you can optimize your use of AWS while keeping costs under control.
Key Points
- Always stop or delete notebook instances when not in use to avoid charges.
- Regularly clean up unused S3 buckets and objects to save on storage costs.
- Monitor your expenses through the AWS Billing Dashboard and set up alerts.
- Use tags (set up earlier in the workshop) to track and monitor costs by resource.
- Following best practices for AWS resource management can significantly reduce costs and improve efficiency.