Convert from .csv to a Shapefile in R
Overview
Teaching: 40 min
Exercises: 20 minQuestions
How can I import CSV files as shapefiles in R?
Objectives
Import .csv files containing x,y coordinate locations into R as a data frame.
Convert a data frame to a spatial object.
Export a spatial object to a text file.
Things You’ll Need To Complete This Episode
See the lesson homepage for detailed information about the software, data, and other prerequisites you will need to work through the examples in this episode.
This episode will introduce how to import spatial points stored in .csv (Comma Separated Value) format into R as an sf spatial object.
We will continue to work with specific packages for working with spatial data. Let’s load all of the packages we have been working with in this lesson.
#library(rgdal)
library(ggplot2)
library(dplyr)
library(sf)
We will also continue to work with the HARV vector data. You may have these layers already loaded into your enviroment, but if not, load them now.
# Learners will have this data loaded from earlier episodes
aoi_boundary_HARV <- st_read("data/vector/HarClip_UTMZ18.shp")
lines_HARV <- st_read("data/vector/HARV_roads.shp")
point_HARV <- st_read("data/vector/HARVtower_UTM18N.shp")
Spatial Data in Text Format
Spatial data are sometimes stored in a text file format (.txt or .csv). The data we downloaded includes a text file called HARV_PlotLocations.csv. This file contains x, y (point) locations for study plot where NEON collects data on
vegetation and other ecological metrics.
While it is very common to store vector data as an Esri shapefile format and most programs can create and read that format, you may encounter data stored in a text file. Text files can be opened in any program and can be converted to any type of vector file format, so it is often useful for long-term storate and sharing with others who may not be familiar with the shapefile format.
We would like to:
- Import the
.csvfile and convert to a spatial object. - Create a map of the plot locations.
- Export the data in a
shapefileformat to share with our colleagues. This shapefile can be imported into any GIS software.
If the text file has an associated x and y location column, then we can
convert it into an sf spatial object. The sf object allows us to store both the x,y values that represent the coordinate location of each point and the associated attribute data - or columns describing each feature in the spatial object.
Import .csv
To begin let’s import the HARV_PlotLoations.csv file with the locations at the NEON Harvard Forest Field Site. Once we import the file, we can check to see if the file contains plot coordinate as x, y attributes.
plot_locations_HARV <- read.csv("data/HARV_PlotLocations.csv")
str(plot_locations_HARV)
'data.frame': 21 obs. of 16 variables:
$ easting : num 731405 731934 731754 731724 732125 ...
$ northing : num 4713456 4713415 4713115 4713595 4713846 ...
$ geodeticDa: chr "WGS84" "WGS84" "WGS84" "WGS84" ...
$ utmZone : chr "18N" "18N" "18N" "18N" ...
$ plotID : chr "HARV_015" "HARV_033" "HARV_034" "HARV_035" ...
$ stateProvi: chr "MA" "MA" "MA" "MA" ...
$ county : chr "Worcester" "Worcester" "Worcester" "Worcester" ...
$ domainName: chr "Northeast" "Northeast" "Northeast" "Northeast" ...
$ domainID : chr "D01" "D01" "D01" "D01" ...
$ siteID : chr "HARV" "HARV" "HARV" "HARV" ...
$ plotType : chr "distributed" "tower" "tower" "tower" ...
$ subtype : chr "basePlot" "basePlot" "basePlot" "basePlot" ...
$ plotSize : int 1600 1600 1600 1600 1600 1600 1600 1600 1600 1600 ...
$ elevation : num 332 342 348 334 353 ...
$ soilTypeOr: chr "Inceptisols" "Inceptisols" "Inceptisols" "Histosols" ...
$ plotdim_m : int 40 40 40 40 40 40 40 40 40 40 ...
We now have a data frame that contains 21 locations (rows) and 16 variables (attributes). Note that all of our character data was imported into R as factor (categorical) data. Next, let’s explore the dataframe to determine whether it contains columns with coordinate values. If we are lucky, our .csv will contain columns with the X and Y locations. We can look for attribute names such as:
- “X” and “Y” OR
- Latitude and Longitude OR
- easting and northing (UTM coordinates)
Let’s check out the column names of our dataframe.
names(plot_locations_HARV)
[1] "easting" "northing" "geodeticDa" "utmZone" "plotID"
[6] "stateProvi" "county" "domainName" "domainID" "siteID"
[11] "plotType" "subtype" "plotSize" "elevation" "soilTypeOr"
[16] "plotdim_m"
Identify X,Y Location Columns
Our column names include several fields that might contain spatial information. The plot_locations_HARV$easting and plot_locations_HARV$northing columns contain coordinate values. We can confirm this by looking at the first six rows of our data.
head(plot_locations_HARV$easting)
[1] 731405.3 731934.3 731754.3 731724.3 732125.3 731634.3
head(plot_locations_HARV$northing)
[1] 4713456 4713415 4713115 4713595 4713846 4713295
We have coordinate values in our data frame. In order to convert our data frame to an sf object, we also need to know the CRS associated with those coordinate values.
There are several ways to figure out the CRS of spatial data in text format.
- We can check the file metadata in hopes that the CRS was recorded in the data.
- We can explore the file itself to see if CRS information is embedded in the file header or somewhere in the data columns.
Following the easting and northing columns, there is a geodeticDa and a
utmZone column. These appear to contain CRS information
(datum and projection). Let’s view those next.
head(plot_locations_HARV$geodeticDa)
[1] "WGS84" "WGS84" "WGS84" "WGS84" "WGS84" "WGS84"
head(plot_locations_HARV$utmZone)
[1] "18N" "18N" "18N" "18N" "18N" "18N"
It is not typical to store CRS information in a column. But this particular
file contains CRS information this way. The geodeticDa and utmZone columns
contain the information that helps us determine the CRS:
geodeticDa: WGS84 – this is geodetic datum WGS84utmZone: 18
In
When Vector Data Don’t Line Up - Handling Spatial Projection & CRS in R
we learned about the components of a proj4 string. We need to know the datum, the projection, and in some cases a zone. For these data we know the datum is WGS84. While the projection is not specified as a column, we are given information about a UTM Zone. This means that our projection is UTM. Therefore, we have everything we need
to assign a CRS to our data frame.
To create the proj4 associated with UTM Zone 18 WGS84 we can look up the
projection on the Spatial Reference website, which contains a list of CRS formats for each projection. From here, we can extract the proj4 string for UTM Zone 18N WGS84.
However, if we have other data in the UTM Zone 18N projection, it’s much
easier to use the st_crs() function to extract the CRS in proj4 format from that object and assign it to our new spatial object.
We’ve seen this CRS before with our Harvard Forest study site (point_HARV).
st_crs(point_HARV)
Coordinate Reference System:
User input: 32618
wkt:
PROJCS["WGS_1984_UTM_Zone_18N",
GEOGCS["GCS_WGS_1984",
DATUM["WGS_1984",
SPHEROID["WGS_84",6378137,298.257223563]],
PRIMEM["Greenwich",0],
UNIT["Degree",0.017453292519943295],
AUTHORITY["EPSG","4326"]],
PROJECTION["Transverse_Mercator"],
PARAMETER["latitude_of_origin",0],
PARAMETER["central_meridian",-75],
PARAMETER["scale_factor",0.9996],
PARAMETER["false_easting",500000],
PARAMETER["false_northing",0],
UNIT["Meter",1],
AUTHORITY["EPSG","32618"]]
The output above shows that the points shapefile is in
UTM zone 18N. We can thus use the CRS from that spatial object to convert our
non-spatial dataframe into an sf object.
Next, let’s create a crs object that we can use to define the CRS of our
sf object when we create it.
utm18nCRS <- st_crs(point_HARV)
utm18nCRS
Coordinate Reference System:
User input: 32618
wkt:
PROJCS["WGS_1984_UTM_Zone_18N",
GEOGCS["GCS_WGS_1984",
DATUM["WGS_1984",
SPHEROID["WGS_84",6378137,298.257223563]],
PRIMEM["Greenwich",0],
UNIT["Degree",0.017453292519943295],
AUTHORITY["EPSG","4326"]],
PROJECTION["Transverse_Mercator"],
PARAMETER["latitude_of_origin",0],
PARAMETER["central_meridian",-75],
PARAMETER["scale_factor",0.9996],
PARAMETER["false_easting",500000],
PARAMETER["false_northing",0],
UNIT["Meter",1],
AUTHORITY["EPSG","32618"]]
class(utm18nCRS)
[1] "crs"
.csv to sf object
Next, let’s convert our dataframe into an sf object. To do this, we need to specify:
- The columns containing X (
easting) and Y (northing) coordinate values - The CRS that the column coordinate represent (units are included in the CRS) - stored in our
utmCRSobject.
We will use the st_as_sf() function to perform the conversion. This function requires the layer to convert, the X and Y locations, and the projection information.
plot_locations_sp_HARV <- st_as_sf(plot_locations_HARV, coords = c("easting", "northing"), crs = utm18nCRS)
We should double check the CRS to make sure it is correct.
st_crs(plot_locations_sp_HARV)
Coordinate Reference System:
User input: 32618
wkt:
PROJCS["WGS_1984_UTM_Zone_18N",
GEOGCS["GCS_WGS_1984",
DATUM["WGS_1984",
SPHEROID["WGS_84",6378137,298.257223563]],
PRIMEM["Greenwich",0],
UNIT["Degree",0.017453292519943295],
AUTHORITY["EPSG","4326"]],
PROJECTION["Transverse_Mercator"],
PARAMETER["latitude_of_origin",0],
PARAMETER["central_meridian",-75],
PARAMETER["scale_factor",0.9996],
PARAMETER["false_easting",500000],
PARAMETER["false_northing",0],
UNIT["Meter",1],
AUTHORITY["EPSG","32618"]]
class(plot_locations_sp_HARV)
[1] "sf" "data.frame"
Plot Spatial Object
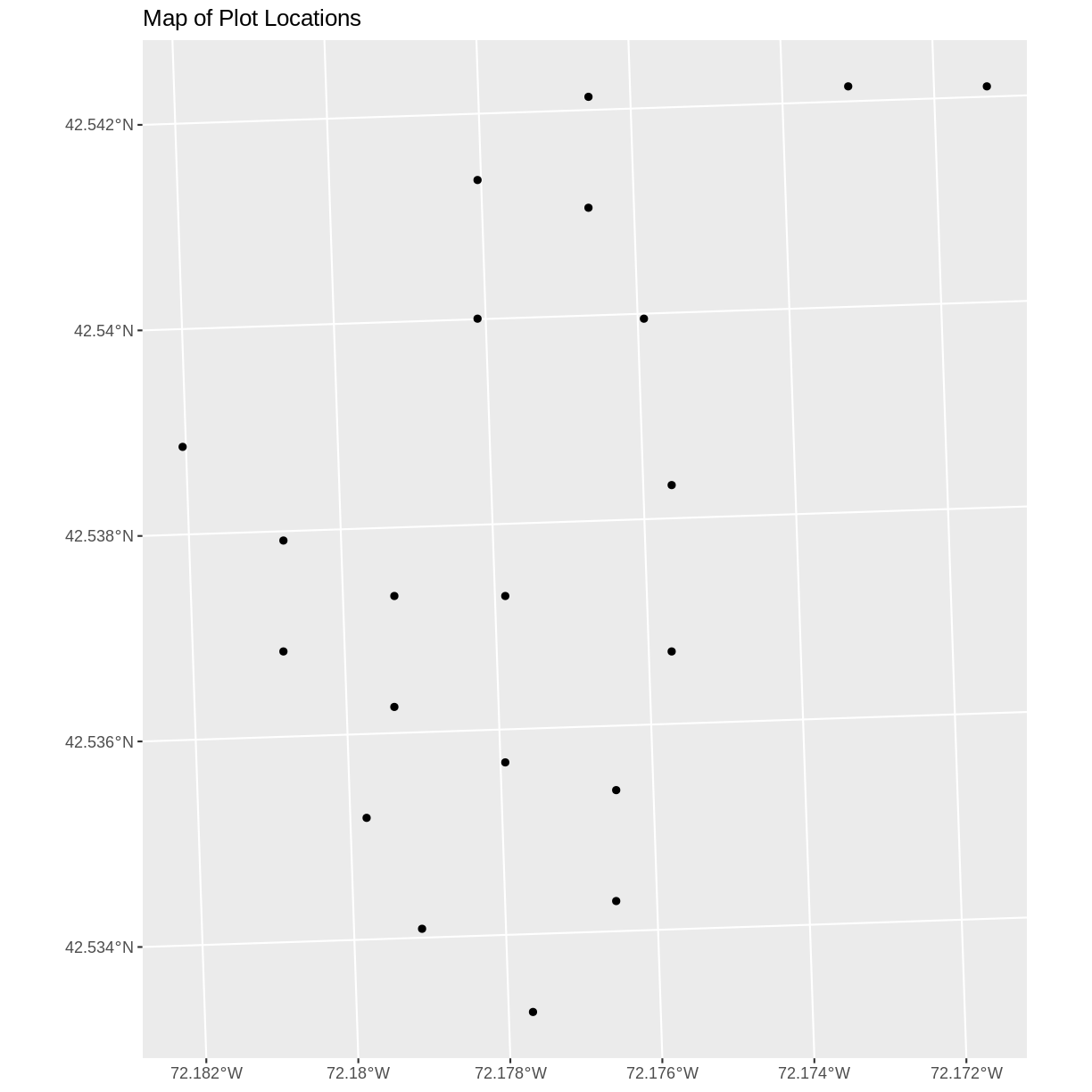
We now have a spatial R object, we can plot our newly created spatial object.
ggplot() +
geom_sf(data = plot_locations_sp_HARV) +
ggtitle("Map of Plot Locations")
Plot Extent
In
Open and Plot Shapefiles in R
we learned about spatial object extent. When we plot several spatial layers in
R using ggplot, all of the layers of the plot are considered in setting the boundaries
of the plot. To show this, let’s plot our aoi_boundary_HARV object with our vegetation plots.
ggplot() +
geom_sf(data = aoi_boundary_HARV) +
geom_sf(data = plot_locations_sp_HARV) +
ggtitle("AOI Boundary Plot")
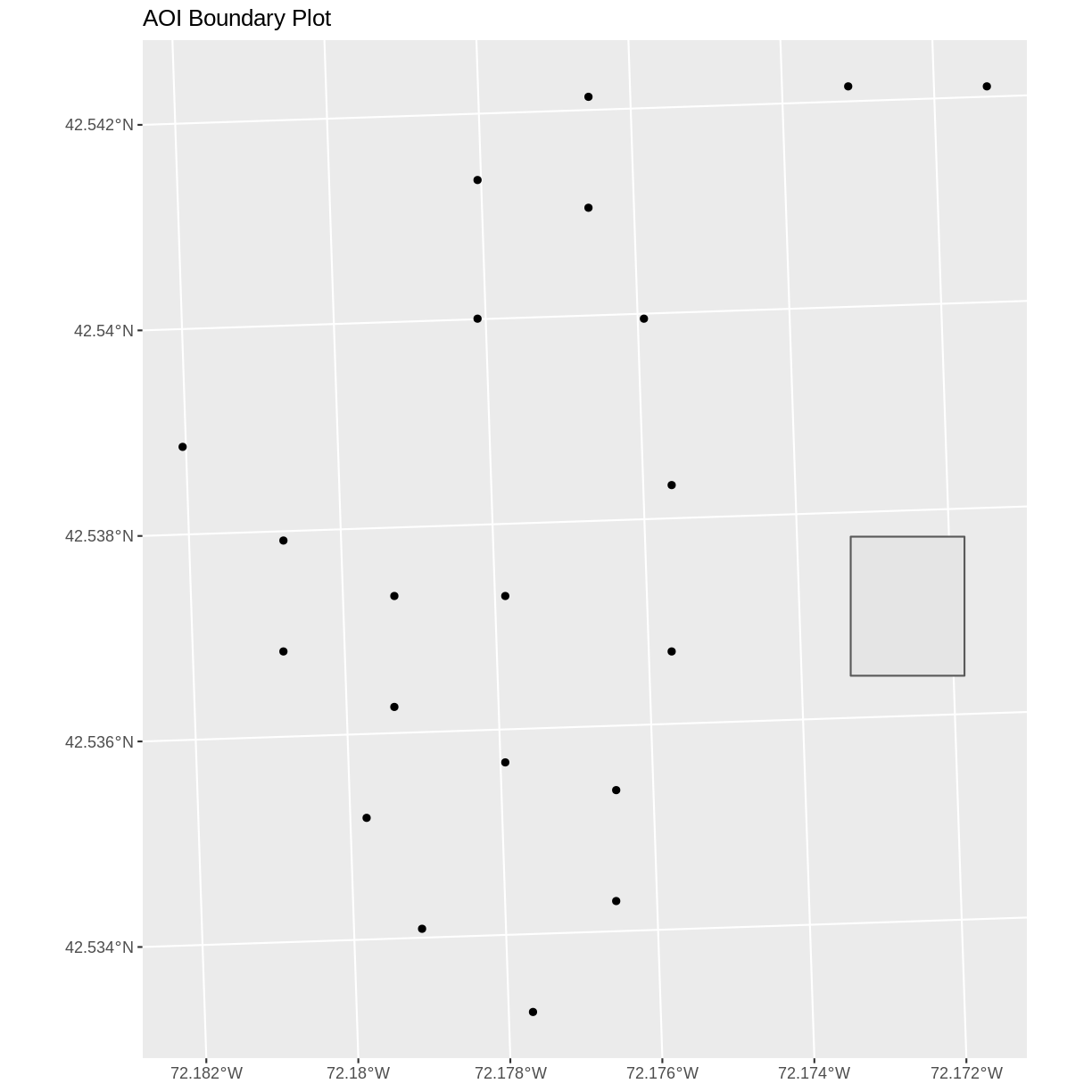
When we plot the two layers together, ggplot sets the plot boundaries
so that they are large enough to include all of the data included in all of the layers.
That’s really handy!
Export a Shapefile
We can write an R spatial object to a shapefile using the st_write function
in sf. To do this we need the following arguments:
- the name of the spatial object (
plot_locations_sp_HARV) - the directory where we want to save our shapefile
(to use
current = getwd()or you can specify a different path) - the name of the new shapefile (
PlotLocations_HARV) - the driver which specifies the file format (ESRI Shapefile)
We can now export the spatial object as a shapefile.
st_write(plot_locations_sp_HARV,
"data/PlotLocations_HARV.shp", driver = "ESRI Shapefile")
Challenge - Import & Plot Additional Points
We want to add two phenology plots to our existing map of vegetation plot locations.
Import the .csv:
HARV/HARV_2NewPhenPlots.csvinto R and do the following:
- Find the X and Y coordinate locations. Which value is X and which value is Y?
- These data were collected in a geographic coordinate system (WGS84). Convert the dataframe into an
sfobject.- Plot the new points with the plot location points from above. Be sure to add a legend. Use a different symbol for the 2 new points!
If you have extra time, feel free to add roads and other layers to your map!
Answers
1) First we will read in the new csv file and look at the data structure.
newplot_locations_HARV <- read.csv("data/HARV_2NewPhenPlots.csv") str(newplot_locations_HARV)'data.frame': 2 obs. of 13 variables: $ decimalLat: num 42.5 42.5 $ decimalLon: num -72.2 -72.2 $ country : chr "unitedStates" "unitedStates" $ stateProvi: chr "MA" "MA" $ county : chr "Worcester" "Worcester" $ domainName: chr "Northeast" "Northeast" $ domainID : chr "D01" "D01" $ siteID : chr "HARV" "HARV" $ plotType : chr "tower" "tower" $ subtype : chr "phenology" "phenology" $ plotSize : int 40000 40000 $ plotDimens: chr "200m x 200m" "200m x 200m" $ elevation : num 358 3462) The US boundary data we worked with previously is in a geographic WGS84 CRS. We can use that data to establish a CRS for this data. First we will extract the CRS from the
country_boundary_USobject and confirm that it is WGS84.country_boundary_US <- st_read("data/vector/US-Boundary-Dissolved-States.shp")Reading layer `US-Boundary-Dissolved-States' from data source `/home/travis/build/UW-Madison-DataScience/r-raster-vector-geospatial/_episodes_rmd/data/vector/US-Boundary-Dissolved-States.shp' using driver `ESRI Shapefile' Simple feature collection with 1 feature and 9 fields geometry type: MULTIPOLYGON dimension: XYZ bbox: xmin: -124.7258 ymin: 24.49813 xmax: -66.9499 ymax: 49.38436 z_range: zmin: 0 zmax: 0 CRS: 4326geogCRS <- st_crs(country_boundary_US) geogCRSCoordinate Reference System: User input: 4326 wkt: GEOGCS["GCS_WGS_1984", DATUM["WGS_1984", SPHEROID["WGS_84",6378137,298.257223563]], PRIMEM["Greenwich",0], UNIT["Degree",0.017453292519943295], AUTHORITY["EPSG","4326"]]Then we will convert our new data to a spatial dataframe, using the
geogCRSobject as our CRS.newPlot.Sp.HARV <- st_as_sf(newplot_locations_HARV, coords = c("decimalLon", "decimalLat"), crs = geogCRS)Next we’ll confirm that the CRS for our new object is correct.
st_crs(newPlot.Sp.HARV)Coordinate Reference System: User input: 4326 wkt: GEOGCS["GCS_WGS_1984", DATUM["WGS_1984", SPHEROID["WGS_84",6378137,298.257223563]], PRIMEM["Greenwich",0], UNIT["Degree",0.017453292519943295], AUTHORITY["EPSG","4326"]]We will be adding these new data points to the plot we created before. The data for the earlier plot was in UTM. Since we’re using
ggplot, it will reproject the data for us.3) Now we can create our plot.
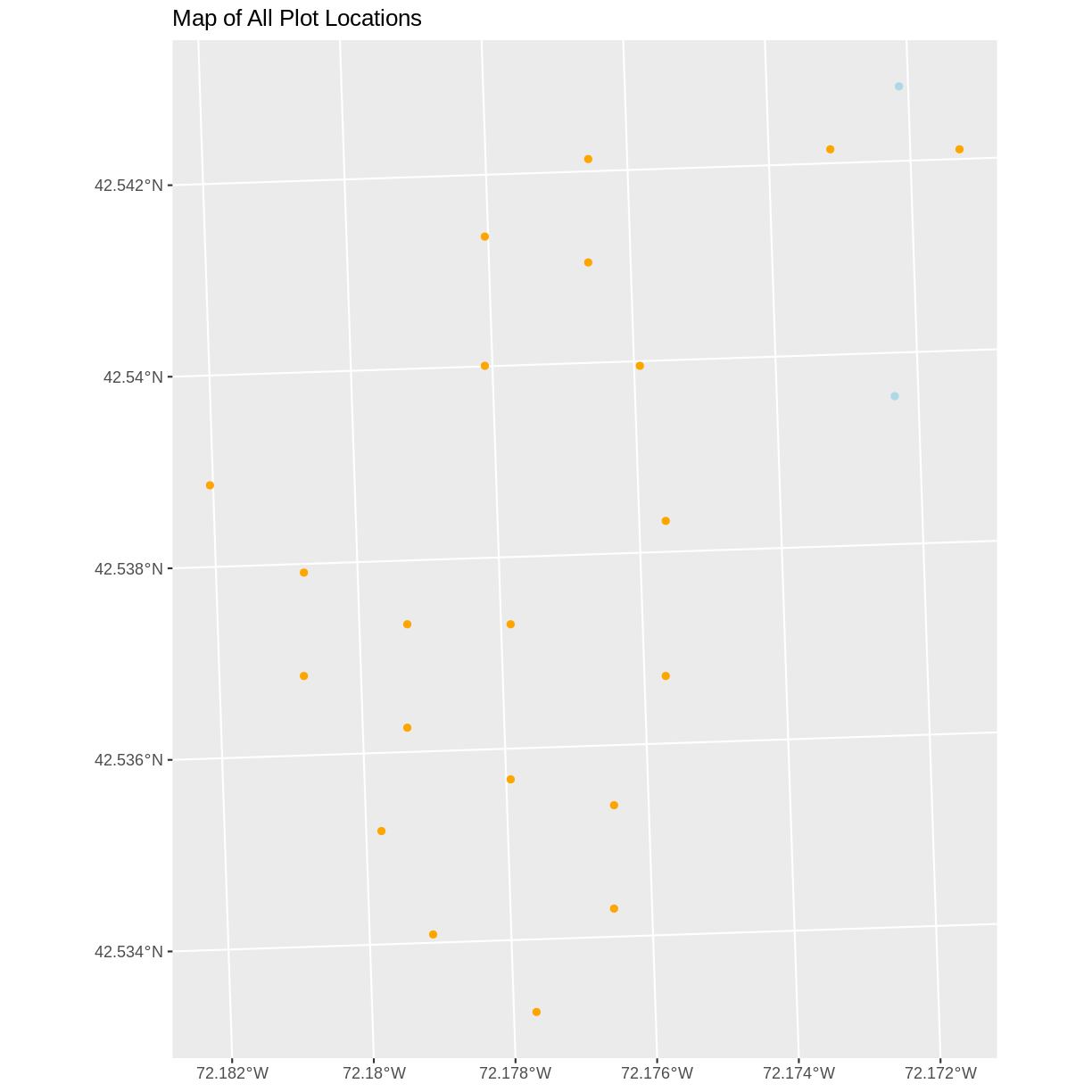
ggplot() + geom_sf(data = plot_locations_sp_HARV, color = "orange") + geom_sf(data = newPlot.Sp.HARV, color = "lightblue") + ggtitle("Map of All Plot Locations")
Key Points
Know the projection (if any) of your point data prior to converting to a spatial object.
Convert a data frame to an
sfobject using thest_as_sf()function.Export an
sfobject as text using thest_write()function.