Day 5: Manipulating Raster Data
Overview
Teaching: min
Exercises: minQuestions
How can I summarize raster data based on vector locations
Objectives
Import .csv files containing x,y coordinate locations and convert to a spatial object
Crop a raster to the extent of a vector layer
Extract values from a raster that correspond to a vector file overlay
Outline
- Addressing Minute questions (10-15 minutes)
- Capstone lesson (30-40 minutes)
- Revisit your spatial question
- Load libraries and import data
- Crop raster with a vector
- Summarize raster data by vector locations
- Keypoints and final questions (10-15 minutes)
1) Revisit your spatial question
On day 1 we brainstormed questions you could ask with geospatial data. You thought about questions you can answer with raster data, and separately, questions you could answer with vector data. Through this workshop you have practiced working with spatial data and have the tools to answer your question.
In this final day we will implement tools you learned for combining raster and vector data and the types of questions you can answer when doing so.
A spatial question may look like the following:
- Where does something occur?
- How does something change across space or time?
- What are the differences between locations?
Examples of spatial questions are:
- What county has the highest recorded precipitation?
- Has mean annual temperature increased in the state?
- How does the precipitation vary between Milwaukee and Madison?
Capstone data question: Does it get hotter in Milwaukee or Madison?
In this capstone lesson we will use the following capstone data:
- World Climate raster of maximum temperature (will download)
- Natural Earth populated places vector point data for cities in Wisconsin (provided
ne_10m_populated_places_Wisconsin.csvfile)
We will use the vector data to crop the raster data to the extent of Wisconsin and summarize the raster data using the point locations of Madison and Milwaukee.
If you are using your own data, please have raster and vector data that cover a similar geographic area. Since we are cropping the raster data by the vector data, it will work best if your vector data is of a smaller geographic area than the raster data.
2) Load libraries and import data
For raster data, we use functions in the raster package. For vector data, you will use functions in the sf package. In addition, because we will visualize our data, we will use the plotting package ggplot2.
Solution
library(raster) library(sf) library(ggplot2)
Load raster data
The capstone data includes the World Climate Data or “WorldClim”. We will use your coding skills to access the worldclim dataset from the raster package, crop it to our extent of interest, and summarize the data. By the end of this section you should have your raster data as a ‘Formal class RasterLayer’ in your Global Environment
World Climate Data
You can learn about and download the WorldClim from the website: https://www.worldclim.org/data/worldclim21.html For an example of how to use the
rasterpackage to access the WorldClim (raster) data and GADM (vector) data, visit this tutorial: https://www.gis-blog.com/r-raster-data-acquisition/
For the WorldClim data, we need to specify the data variable and the spatial resolution. We will use the [“bio” variable] (https://www.worldclim.org/data/bioclim.html), which are 19 historial bioclimatic variables. The bio variables include things like:
- annual mean temperature
- max temperataure of the warmest month
- annual precipitation
- precipiation of wettest month
The bio variables are available at spatial resolutions of 10 minutes, 5 minutes, 2.5 minutes, and 30 seconds.
Wait, why is spatial resolution in units of time? This is because the spatial coordinates on a sphere are often expressed as degrees-minutes-seconds. 10 minutes is the biggest and 30 seconds is the smallest. An online conversion tool is helpful to understand these unit conversions(https://www.opendem.info/arc2meters.html)
For the capstone data, we want to use the largest spatial resolution of 10 minutes. This means each pixel represents about 13 km on the ground at the approximate latitude of Wisconsin. Within the “bio” variables, we will use the “bio5” variable that represents the maxiumum temperature of the warmest month.
Solution
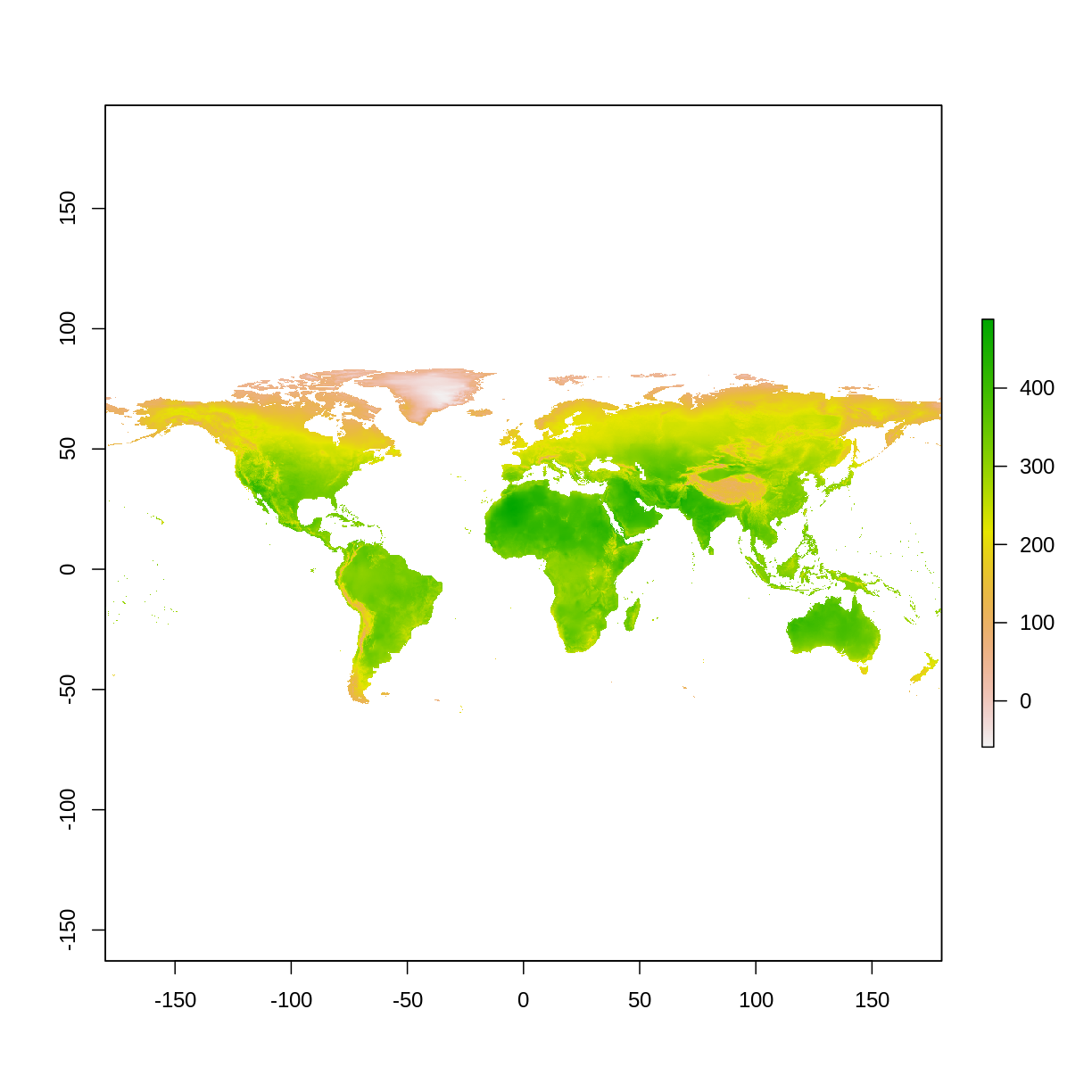
# get worldclim data, variable "bio" with a 10 minute resolution # this line is downloading data # if you selected a resolution higher than 10 (with a res of 0.5,2.5, or 5) it may take a while to download worldClim_bio_global <- getData("worldclim", var="bio", res=10) # use only the bio5 variable (BIO5 = Max Temperature of Warmest Month maxTemp_global <- worldClim_bio_global$bio5 # plot data plot(maxTemp_global)
Load vector data
The capstone data includes the Natural Earth Populated Places dataset. We have provided the dataset for you as a “.csv” file in the Capstone data folder. By the end of this section you should have a simple feature (‘sf’) data frame type of object in your Global Environment.
Natural Earth Data
You can learn about and download the Natural Earth dataset from the website: https://www.naturalearthdata.com/ You can also download Natural Earth data directly into R using the [
rnaturalearthpackage] (https://cran.r-project.org/web/packages/rnaturalearth/README.html)
Solution
# import point data from a csv file WIcities <- read.csv("../data/ne_10m_populated_places_Wisconsin.csv")Warning in file(file, "rt"): cannot open file '../data/ ne_10m_populated_places_Wisconsin.csv': No such file or directoryError in file(file, "rt"): cannot open the connection# check out the data colnames(WIcities)Error in is.data.frame(x): object 'WIcities' not found
Recall from your lesson how to import point data and convert it to a spatial data layer in R.
When looking at the column names we see that there is a lot of data! To convert to a spatial points data layer, we need to know information about the coordinate system and the coordinates for each point.
The column names LATITUDE and LONGITUDE have what we need!
Solution
# convert data frame to sf type of object by specifying coordinates (as xy) and the projection as a crs WIcities_points <- st_as_sf(WIcities, coords=c('LONGITUDE','LATITUDE'), crs=crs(maxTemp_global))Error in st_as_sf(WIcities, coords = c("LONGITUDE", "LATITUDE"), crs = crs(maxTemp_global)): object 'WIcities' not found# check class of object - should be "sf" data frame class(WIcities_points)Error in eval(expr, envir, enclos): object 'WIcities_points' not found
A critical part of working with raster and vector data together is that the datasets are in the same coordinate system.
Does it make sense? Are they the same? You can use the functions in the raster package to check out your data.
crs(maxTemp_global)
CRS arguments: +proj=longlat +datum=WGS84 +no_defs
crs(WIcities_points)
Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'crs': object 'WIcities_points' not found
extent(maxTemp_global)
class : Extent
xmin : -180
xmax : 180
ymin : -60
ymax : 90
extent(WIcities_points)
Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'extent': object 'WIcities_points' not found
res(maxTemp_global)
[1] 0.1666667 0.1666667
Finally, let’s subset our spatial data to only include the city points we are interested in. For the sample dataset, we are going to subset the data to only include the cities of Madison and Milwaukee.
If you are using your own data, you may or may not need to do this subset or you might want to choose different locations.
Solution
WIcities_points$NAMEError in eval(expr, envir, enclos): object 'WIcities_points' not foundMM_points <- WIcities_points[WIcities_points$NAME=='Madison'|WIcities_points$NAME=='Milwaukee',]Error in eval(expr, envir, enclos): object 'WIcities_points' not found
3) Crop raster with vector
It may be useful to limit the raster data to only areas of interest. This is helpful when you load a larger raster file, such as data for the entire globe, but are only interested in a small part. For our capstone data, we loaded the global temperature data, but we are only interested in the data for Wisconsin, and specifically, the data for Milwaukee and Madison. Let’s use the crop() function in the raster package to cut out the maximum temperature data to only the area for the Wisconsin cities.
Solution
maxTemp_WI <- crop(x = maxTemp_global, y = extent(WIcities_points),snap="out")Error in .local(x, y, ...): Cannot get an Extent object from argument ymaxTemp_WI_df <- as.data.frame(maxTemp_WI, xy=T)Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'as.data.frame': object 'maxTemp_WI' not foundggplot() + geom_raster(data = maxTemp_WI_df,aes(x = x, y = y, fill = bio5)) + geom_sf(data = MM_points,color="yellow",size=5) + coord_sf()Error in fortify(data): object 'maxTemp_WI_df' not found
4) Summarize raster data by vector locations
Now that we have our raster layer that contains the data we want to summarize, and the vector data that contains the locations indicating where we want to summarize the data, we can now put them together.
If you are working with your own data, ask yourself:
- How do I want to summarize the data? For example, do I want a single mean value? A maximum? A distribution of all values?
- What is the area I want to summarize? If it’s a point, do you want a certain distance around the point? Or, if it’s an area, you will include all of the raster values within that area.
Extract pixel values
For the capstone data, we want to extract all the pixel values within a distance buffer of the two city points. Since our data is “unprojected” and therefore the units are in latitude and longitude, the buffer distance will be in meters (hint: this information is in the Help file for the extract() function!)
Geographic and projected unit reminder
Remember, your raster is “unprojected” meaning that the pixels represent distances on a sphere of degrees, minutes, and seconds. You’ll need to consider is the conversation from degrees to meters. The conversions depends on where you are on the globe. For example, at the equator, 1 degree is 111 km. This distance decreases as you move closer to the poles. Try setting the buffer to 50 km or 50,000 meters. This is something you can play around with as you explore the data.
Solution
# extract pixel values from the raster based on the Milwaukee and Madison points # using a buffer size of 50,000 (50 km around the each point) because the units are in meters maxTemp_MM <- extract(x=maxTemp_global,df=TRUE, y=MM_points, buffer=50000)Error in h(simpleError(msg, call)): error in evaluating the argument 'y' in selecting a method for function 'extract': object 'MM_points' not found
Convert data and plot
The last few steps will be small conversions of the datatypes to make it easy for summarizing and plotting.
Solution
# convert to data frame for each calculations and plotting df_maxTemp_MM <- as.data.frame(maxTemp_MM)Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'as.data.frame': object 'maxTemp_MM' not found# clean up data frame because we want to give IDs names and convert the temperature data df_maxTemp_MM$ID <- as.factor(df_maxTemp_MM$ID)Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'as.factor': object 'df_maxTemp_MM' not foundlevels(df_maxTemp_MM$ID) = levels(as.factor(MM_points$NAME))Error in h(simpleError(msg, call)): error in evaluating the argument 'x' in selecting a method for function 'levels': error in evaluating the argument 'x' in selecting a method for function 'as.factor': object 'MM_points' not founddf_maxTemp_MM$bio5 <- df_maxTemp_MM$bio5/10Error in eval(expr, envir, enclos): object 'df_maxTemp_MM' not found# plot data ggplot(data = df_maxTemp_MM, aes(x = bio5,fill=ID)) + geom_histogram() + ggtitle("Histogram of maximum temperatures within 50 km of the city") + xlab("Maximum temperature (C)") + ylab("Frequency of Pixels")Error in ggplot(data = df_maxTemp_MM, aes(x = bio5, fill = ID)): object 'df_maxTemp_MM' not found
The graph above shows us the distribution of maximum temperatures within 50 km of Madison and Milwaukee. We have a distribution of values because the extracted data represents each individual pixel within the 50 km buffer. This graph shows us that the maximum temperatures recorded in the Madison are tend to be higher than the maximum temperatures recorded in the Milwaukee region. Does this pattern make sense with your knowledge of the area and the data? Remember, visualizing the data can be important in understanding and explaining patterns.
Key Points
Convert a data frame to an
sfobject using thest_as_sf()function.Use the
crop()andextract()functions to get raster data of spatial subsets.Remember to check if your data align spatially by looking at the projection, extent, and units